Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis
- PMID: 18081427
- PMCID: PMC2098807
- DOI: 10.1371/journal.pgen.0030214
Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis
Abstract
A novel candidate metastasis modifier, ribosomal RNA processing 1 homolog B (Rrp1b), was identified through two independent approaches. First, yeast two-hybrid, immunoprecipitation, and functional assays demonstrated a physical and functional interaction between Rrp1b and the previous identified metastasis modifier Sipa1. In parallel, using mouse and human metastasis gene expression data it was observed that extracellular matrix (ECM) genes are common components of metastasis predictive signatures, suggesting that ECM genes are either important markers or causal factors in metastasis. To investigate the relationship between ECM genes and poor prognosis in breast cancer, expression quantitative trait locus analysis of polyoma middle-T transgene-induced mammary tumor was performed. ECM gene expression was found to be consistently associated with Rrp1b expression. In vitro expression of Rrp1b significantly altered ECM gene expression, tumor growth, and dissemination in metastasis assays. Furthermore, a gene signature induced by ectopic expression of Rrp1b in tumor cells predicted survival in a human breast cancer gene expression dataset. Finally, constitutional polymorphism within RRP1B was found to be significantly associated with tumor progression in two independent breast cancer cohorts. These data suggest that RRP1B may be a novel susceptibility gene for breast cancer progression and metastasis.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
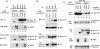

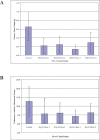
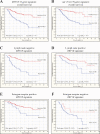


References
-
- Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, et al. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. - PubMed
-
- Guarneri V, Conte PF. The curability of breast cancer and the treatment of advanced disease. Eur J Nucl Med Mol Imaging. 2004;31(Suppl 1):S149–S161. - PubMed
-
- Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–644. - PubMed
-
- Hunter KW, Broman KW, Voyer TL, Lukes L, Cozma D, et al. Predisposition to efficient mammary tumor metastatic progression is linked to the breast cancer metastasis suppressor gene Brms1. Cancer Res. 2001;61:8866–8872. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

