Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H
- PMID: 18081690
- PMCID: PMC2276932
- DOI: 10.1111/j.1365-2249.2007.03552.x
Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H
Abstract
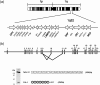
Factor H is an abundant plasma glycoprotein that plays a critical role in the regulation of the complement system in plasma and in the protection of host cells and tissues from damage by complement activation. Several recent studies have described the association of genetic variations of the complement factor H gene (CFH) with atypical haemolytic uraemic syndrome (aHUS), age-related macular degeneration (AMD) and membranoproliferative glomerulonephritis (MPGN). This review summarizes our current knowledge of CFH genetics and examines the CFH genotype-phenotype correlations that are helping to understand the molecular basis underlying these renal and ocular pathologies.
Figures
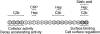



References
-
- Law SKA, Reid KBM. Complement. 2. Oxford: IRL Press; 1995.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

