An analysis of the epitope knowledge related to Mycobacteria
- PMID: 18081934
- PMCID: PMC2228276
- DOI: 10.1186/1745-7580-3-10
An analysis of the epitope knowledge related to Mycobacteria
Abstract
Background: Tuberculosis, caused by the bacterium Mycobacterium tuberculosis, remains a leading cause of infectious disease morbidity and mortality, and is responsible for more than 2 million deaths a year. Reports about extremely drug resistant (XDR) strains have further heightened the sense of urgency for the development of novel strategies to prevent and treat TB. Detailed knowledge of the epitopes recognized by immune responses can aid in vaccine and diagnostics development, and provides important tools for basic research. The analysis of epitope data corresponding to M. tuberculosis can also identify gaps in our knowledge, and suggest potential areas for further research and discovery. The Immune Epitope Database (IEDB) is compiled mainly from literature sources, and describes a broad array of source organisms, including M. tuberculosis and other Mycobacterial species.
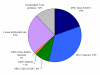
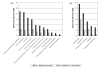
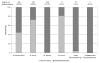
Description: A comprehensive analysis of IEDB data regarding the genus Mycobacteria was performed. The distribution of antibody/B cell and T cell epitopes was analyzed in terms of their associated recognition cell type effector function and chemical properties. The various species, strains and proteins which the epitope were derived, were also examined. Additional variables considered were the host in which the epitopes were defined, the specific TB disease state associated with epitope recognition, and the HLA associated with disease susceptibility and endemic regions were also scrutinized. Finally, based on these results, standardized reference datasets of mycobacterial epitopes were generated.
Conclusion: All current TB-related epitope data was cataloged for the first time from the published literature. The resulting inventory of more than a thousand different epitopes should prove a useful tool for the broad scientific community. Knowledge gaps specific to TB epitope data were also identified. In summary, few non-peptidic or post-translationally modified epitopes have been defined. Most importantly epitopes have apparently been defined from only 7% of all ORFs, and the top 30 most frequently studied protein antigens contain 65% of the epitopes, leaving the majority of M. tuberculosis genome unexplored. A lack of information related to the specific strains from which epitopes are derived is also evident. Finally, the generation of reference lists of mycobacterial epitopes should also facilitate future vaccine and diagnostic research.
Figures
References
-
- Immune Epitope Database and Analysis Resource (IEDB) website http://www.immuneepitope.org - PMC - PubMed
-
- Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger SP, Stewart S, Surko P, Way S, Wilson S, Sette A. The design and implementation of the immune epitope database and analysis resource. Immunogenetics. 2005;57:326–336. doi: 10.1007/s00251-005-0803-5. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
