Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly
- PMID: 18082608
- PMCID: PMC2246091
- DOI: 10.1016/j.molcel.2007.09.022
Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly
Abstract
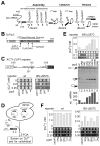
ATPase-facilitated steps during spliceosome function have been postulated to afford opportunities for kinetic proofreading. Spliceosome assembly requires the ATPase Prp5p, whose activity might thus impact fidelity during initial intron recognition. Using alanine mutations in S. cerevisiae Prp5p, we identified a suboptimal intron whose splicing could be improved by altered Prp5p activity and then, using this intron, screened for potent prp5 mutants. These prp5 alleles specifically alter branch region selectivity, with improved splicing in vivo of suboptimal substrates correlating with reduced ATPase activity in vitro for a series of mutants in ATPase motif III (SAT). Because these effects are abrogated by compensatory U2 snRNA mutations or other changes that increase branch region-U2 pairing, these results explicitly link a fidelity event with a defined physical structure, the branch region-U2 snRNA duplex, and provide strong evidence that progression of the splicing pathway requires branch region-U2 snRNA pairing prior to Prp5p-facilitated conformational change.
Figures



References
-
- Abu Dayyeh BK, Quan TK, Castro M, Ruby SW. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J Biol Chem. 2002;277:20221–20233. - PubMed
-
- Burgess S, Couto JR, Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. - PubMed
-
- Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993;18:381–384. - PubMed
-
- Couto JR, Tamm J, Parker R, Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1:445–455. - PubMed
-
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

