Trm9-catalyzed tRNA modifications link translation to the DNA damage response
- PMID: 18082610
- PMCID: PMC2211415
- DOI: 10.1016/j.molcel.2007.09.021
Trm9-catalyzed tRNA modifications link translation to the DNA damage response
Abstract
Transcriptional and posttranslational signals are known mechanisms that promote efficient responses to DNA damage. We have identified Saccharomyces cerevisiae tRNA methyltransferase 9 (Trm9) as an enzyme that prevents cell death via translational enhancement of DNA damage response proteins. Trm9 methylates the uridine wobble base of tRNAARG(UCU) and tRNAGLU(UUC). We used computational and molecular approaches to predict that Trm9 enhances the translation of some transcripts overrepresented with specific arginine and glutamic acid codons. We found that translation elongation factor 3 (YEF3) and the ribonucleotide reductase (RNR1 and RNR3) large subunits are overrepresented with specific arginine and glutamic acid codons, and we demonstrated that Trm9 significantly enhances Yef3, Rnr1, and Rnr3 protein levels. Additionally, we identified 425 genes, which included YEF3, RNR1, and RNR3, with a unique codon usage pattern linked to Trm9. We propose that Trm9-specific tRNA modifications enhance codon-specific translation elongation and promote increased levels of key damage response proteins.
Figures

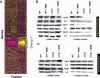
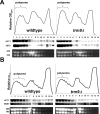

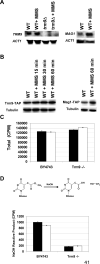

Comment in
-
Better living through biochemistry.Nat Methods. 2011 Feb;8(2):108-9. doi: 10.1038/nmeth0211-108b. Nat Methods. 2011. PMID: 21355118 No abstract available.
References
-
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. - PubMed
-
- Anand M, Chakraburtty K, Marton MJ, Hinnebusch AG, Kinzy TG. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J Biol Chem. 2003;278:6985–6991. - PubMed
-
- Barrows LR, Magee PN. Nonenzymatic methylation of DNA by S-adenosylmethionine in vitro. Carcinogenesis. 1982;3:349–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

