Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance
- PMID: 18083799
- PMCID: PMC2258673
- DOI: 10.1128/JB.01592-07
Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance
Abstract
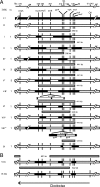
The high frequency of between-strain genetic recombinants of Chlamydia trachomatis among isolates obtained from human sexually transmitted infections suggests that lateral gene transfer (LGT) is an important means by which C. trachomatis generates variants that have enhanced relative fitness. A mechanism for LGT in C. trachomatis has not been described, and investigation of this phenomenon by experimentation has been hampered by the obligate intracellular development of this pathogen. We describe here experiments that readily detected LGT between strains of C. trachomatis in vitro. Host cells were simultaneously infected with an ofloxacin-resistant (Ofx(r)) mutant of a serovar L1 strain (L1:Ofx(r)-1) and a rifampin-resistant (Rif(r)) mutant of a serovar D strain (D:Rif(r)-1). Development occurred in the absence of antibiotics, and the progeny were subjected to selection for Ofx(r) Rif(r) recombinants. The parental strains differed at many polymorphic nucleotide sites, and DNA sequencing was used to map genetic crossovers and to determine the parental sources of DNA segments in 14 recombinants. Depending on the assumed DNA donor, the estimated minimal length of the transferred DNA was > or = 123 kb in one recombinant but was > or = 336 to > or = 790 kb in all other recombinants. Such trans-DNA lengths have been associated only with conjugation in known microbial LGT systems, but natural DNA transformation remains a conceivable mechanism. LGT studies can now be performed with diverse combinations of C. trachomatis strains, and they could have evolutionary interest and yield useful recombinants for functional analysis of allelic differences between strains.
Figures
References
-
- Dean, D. 2002. Pathogenesis of chlamydial ocular infections, p. 1-20. In K. Tasman and E. Jaeger (ed.), Duane's foundations of clinical ophthalmology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

