Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus
- PMID: 18083809
- PMCID: PMC2238224
- DOI: 10.1128/JB.01128-07
Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus
Abstract
Staphylococcus aureus becomes resistant to methicillin by acquiring a genomic island, known as staphylococcal chromosome cassette mec (SCCmec), which contains the methicillin resistance determinant, mecA. SCCmec is site-specifically integrated into the staphylococcal chromosome at a locus known as the SCCmec attachment site (attB). In an effort to gain a better understanding of the potential that methicillin-sensitive S. aureus (MSSA) isolates have for acquiring SCCmec, the nucleotide sequences of attB and surrounding DNA regions were examined in a diverse collection of 42 MSSA isolates. The chromosomal region surrounding attB varied among the isolates studied and appears to be a common insertion point for acquired foreign DNA. Insertions of up to 15.1 kb were found containing open reading frames with homology to enterotoxin genes, restriction-modification systems, transposases, and several sequences that have not been previously described in staphylococci. Two groups, containing eight and four isolates, had sequences found in known SCCmec elements, suggesting SCCmec elements may have evolved through repeated DNA insertions at this locus. In addition, the attB sequences of the majority of MSSA isolates in this collection differ from the attB sequences of strains for which integrase-mediated SCCmec insertion or excision has been demonstrated, suggesting that some S. aureus isolates may lack the ability to site-specifically integrate SCCmec into their chromosomes.
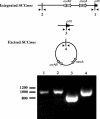
Figures





References
-
- Anonymous. 2004. National Nosocomial Infections Surveillance (NNIS) System Report; data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32470-485. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. - PubMed
-
- Chambers, H. F. 2003. Solving staphylococcal resistance to beta-lactams. Trends Microbiol. 11145-148. - PubMed
-
- Cooper, J. E., and E. J. Feil. 2006. The phylogeny of Staphylococcus aureus—which genes make the best intra-species markers? Microbiology 1521297-1305. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

