Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon
- PMID: 18083811
- PMCID: PMC2238217
- DOI: 10.1128/JB.01764-07
Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon
Abstract
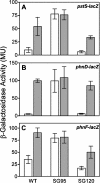
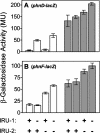
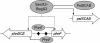
The uptake of phosphate into the cell via high-affinity, phosphate-specific transport systems has been studied with several species of mycobacteria. All of these species have been shown to contain several copies of such transport systems, which are synthesized in response to phosphate limitation. However, the mechanisms leading to the expression of the genes encoding these transporters have not been studied. This study reports on the investigation of the regulation of the pstSCAB and the phnDCE operons of Mycobacterium smegmatis. The phn locus contains an additional gene, phnF, encoding a GntR-like transcriptional regulator. Expression analyses of a phnF deletion mutant demonstrated that PhnF acts as a repressor of the phnDCE operon but does not affect the expression of pstSCAB. The deletion of pstS, which is thought to cause the constitutive expression of genes regulated by the two-component system SenX3-RegX3, led to the constitutive expression of the transcriptional fusions pstS-lacZ, phnD-lacZ, and phnF-lacZ, suggesting that phnDCE and phnF are conceivably new members of the SenX3-RegX3 regulon of M. smegmatis. Two presumptive binding sites for PhnF in the intergenic region between phnD and phnF were identified and shown to be required for the repression of phnD and phnF, respectively. We propose a model in which the transcription of pstSCAB is controlled by the two-component SenX3-RegX3 system, while phnDCE and phnF are subject to dual control by SenX3-RegX3 and PhnF.
Figures





References
-
- Aravind, L., and V. Anantharaman. 2003. HutC/FarR-like bacterial transcription factors of the GntR family contain a small molecule-binding domain of the chorismate lyase fold. FEMS Microbiol. Lett. 22217-23. - PubMed
-
- Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14892-893. - PubMed
-
- Gebhard, S., S. L. Tran, and G. M. Cook. 2006. The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology 1523453-3465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

