Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY
- PMID: 18083822
- PMCID: PMC2238190
- DOI: 10.1128/JB.01327-07
Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY
Abstract
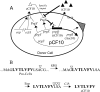
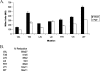
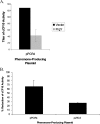
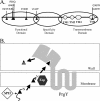
Conjugative transfer of the Enterococcus faecalis plasmid pCF10 is induced by the peptide pheromone cCF10 when recipient-produced cCF10 is detected by donors. cCF10 is produced by proteolytic processing of the signal sequence of a chromosomally encoded lipoprotein (CcfA). In donors, endogenously produced cCF10 is carefully controlled to prevent constitutive expression of conjugation functions, an energetically wasteful process, except in vivo, where endogenous cCF10 induces a conjugation-linked virulence factor. Endogenous cCF10 is controlled by two plasmid-encoded products; a membrane protein PrgY reduces pheromone levels in donors, and a secreted inhibitor peptide iCF10 inhibits the residual endogenous pheromone that escapes PrgY control. In this study we genetically determined the amino acid specificity determinants within PrgY, cCF10, and the cCF10 precursor that are necessary for cCF10 processing and for PrgY-mediated control. We showed that amino acid residues 125 to 241 of PrgY are required for specific recognition of cCF10 and that PrgY recognizes determinants within the heptapeptide cCF10 sequence, supporting a direct interaction between PrgY and mature cCF10. In addition, we found that a regulated intramembrane proteolysis (RIP) family pheromone precursor-processing protein Eep recognizes amino acids N-terminal to cCF10 in the signal sequence of CcfA. These results support a model where Eep directly targets pheromone precursors for RIP and PrgY interacts directly with the mature cCF10 peptide during processing. Despite evidence that both PrgY and Eep associate with cCF10 in or near the membrane, results presented here indicate that these two proteins function independently.
Figures




References
-
- Bae, T., and G. M. Dunny. 2001. Dominant-negative mutants of prgX: evidence for a role for PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 391307-1320. - PubMed
-
- Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315995-1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

