Candida albicans Als adhesins have conserved amyloid-forming sequences
- PMID: 18083824
- PMCID: PMC2394976
- DOI: 10.1128/EC.00309-07
Candida albicans Als adhesins have conserved amyloid-forming sequences
Abstract
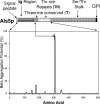
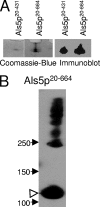
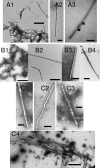
The cell wall-bound Als adhesins of Candida albicans mediate both yeast-to-host tissue adherence and yeast aggregation. This aggregation is amyloid-like, with self-propagating secondary-structure changes, amyloid-characteristic dye binding, and induced birefringence (J. M. Rauceo, N. K. Gaur, K. G. Lee, J. E. Edwards, S. A. Klotz, and P. N. Lipke, Infect. Immun. 72:4948-4955, 2004). Therefore, we determined whether Als proteins could form amyloid fibers with properties like those in cellular aggregation. The beta-aggregation predictor TANGO identified a heptapeptide sequence present in a highly conserved sequence with amyloid-forming potential in Als1p, Als3p, and Als5p. A tridecapeptide containing this sequence formed fibers that bound Congo red and thioflavin T and had characteristic amyloid morphology. Als5p(20-431) and Als5p(20-664), large fragments of Als5p containing the amyloid sequence, also formed amyloid-like fibers and bound Congo red under native conditions. K(a)/K(s) analysis showed that the amyloid-forming sequences are highly conserved in Als proteins and evolve more slowly than other regions of the proteins. Therefore, amyloid-forming ability itself is conserved in these proteins.
Figures

References
-
- Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 43769-87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

