Global analysis of mRNA splicing
- PMID: 18083834
- PMCID: PMC2212243
- DOI: 10.1261/rna.868008
Global analysis of mRNA splicing
Abstract
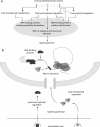
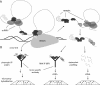
Alternative mRNA splicing is a rich source of transcript diversity in eukaryotic cells with broad roles in development and disease. Systems-wide experimental methods have started to define how global splicing regulation shapes complex biological properties and pathways. Here, we review these approaches, describe recent insights they have yielded, and discuss avenues of future investigation.
Figures

Similar articles
-
Systems analysis of alternative splicing and its regulation.Wiley Interdiscip Rev Syst Biol Med. 2010 Sep-Oct;2(5):550-565. doi: 10.1002/wsbm.84. Wiley Interdiscip Rev Syst Biol Med. 2010. PMID: 20836047 Review.
-
Alternative splicing: new insights from global analyses.Cell. 2006 Jul 14;126(1):37-47. doi: 10.1016/j.cell.2006.06.023. Cell. 2006. PMID: 16839875 Review.
-
Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense?Trends Biochem Sci. 2008 Aug;33(8):385-93. doi: 10.1016/j.tibs.2008.06.001. Epub 2008 Jul 11. Trends Biochem Sci. 2008. PMID: 18621535 Review.
-
Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis.RNA. 2008 Feb;14(2):284-96. doi: 10.1261/rna.725208. Epub 2007 Dec 11. RNA. 2008. PMID: 18073345 Free PMC article.
-
Alternative splicing: a bioinformatics perspective.Mol Biosyst. 2007 Jul;3(7):473-7. doi: 10.1039/b702485c. Epub 2007 May 18. Mol Biosyst. 2007. PMID: 17579772 Review.
Cited by
-
Introns regulate gene expression in Cryptococcus neoformans in a Pab2p dependent pathway.PLoS Genet. 2013;9(8):e1003686. doi: 10.1371/journal.pgen.1003686. Epub 2013 Aug 15. PLoS Genet. 2013. PMID: 23966870 Free PMC article.
-
TDP-43 regulates cancer-associated microRNAs.Protein Cell. 2018 Oct;9(10):848-866. doi: 10.1007/s13238-017-0480-9. Epub 2017 Sep 26. Protein Cell. 2018. PMID: 28952053 Free PMC article.
-
LATENT RANK CHANGE DETECTION FOR ANALYSIS OF SPLICE-JUNCTION MICROARRAYS WITH NONLINEAR EFFECTS.Ann Appl Stat. 2011;5(1):364-380. doi: 10.1214/10-AOAS389SUPP. Ann Appl Stat. 2011. PMID: 23335951 Free PMC article.
-
Expression map of the human exome in CD34+ cells and blood cells: increased alternative splicing in cell motility and immune response genes.PLoS One. 2010 Feb 1;5(2):e8990. doi: 10.1371/journal.pone.0008990. PLoS One. 2010. PMID: 20126548 Free PMC article.
-
Genome-wide RNAi screen discovers functional coupling of alternative splicing and cell cycle control to apoptosis regulation.Cell Cycle. 2010 Nov 15;9(22):4419-21. doi: 10.4161/cc.9.22.14051. Epub 2010 Nov 15. Cell Cycle. 2010. PMID: 21088477 Free PMC article. No abstract available.
References
-
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed
-
- Blanchette, M., Labourier, E., Green, R.E., Brenner, S.E., Rio, D.C. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004;14:775–786. - PubMed
-
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
