Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA
- PMID: 18083837
- PMCID: PMC2212247
- DOI: 10.1261/rna.697308
Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA
Abstract
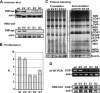
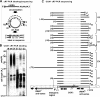
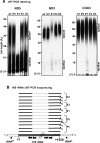
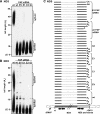
Polynucleotide phosphorylase (PNPase) is a diverse enzyme, involved in RNA polyadenylation, degradation, and processing in prokaryotes and organelles. However, in human mitochondria, PNPase is located in the intermembrane space (IMS), where no mitochondrial RNA (mtRNA) is known to be present. In order to determine the nature and degree of its involvement in mtRNA metabolism, we stably silenced PNPase by establishing HeLa cell lines expressing PNPase short-hairpin RNA (shRNA). Processing and polyadenylation of mt-mRNAs were significantly affected, but, to different degrees in different genes. For instance, the stable poly(A) tails at the 3' ends of COX1 transcripts were abolished, while COX3 poly(A) tails remained unaffected and ND5 and ND3 poly(A) extensions increased in length. Despite the lack of polyadenylation at the 3' end, COX1 mRNA and protein accumulated to normal levels, as was the case for all 13 mt-encoded proteins. Interestingly, ATP depletion also altered poly(A) tail length, demonstrating that adenylation of mtRNA can be manipulated by indirect, environmental means and not solely by direct enzymatic activity. When both PNPase and the mitochondrial poly(A)-polymerase (mtPAP) were concurrently silenced, the mature 3' end of ND3 mRNA lacked poly(A) tails but retained oligo(A) extensions. Furthermore, in mtPAP-silenced cells, truncated adenylated COX1 molecules, considered to be degradation intermediates, were present but harbored significantly shorter tails. Together, these results suggest that an additional mitochondrial polymerase, yet to be identified, is responsible for the oligoadenylation of mtRNA and that PNPase, although located in the IMS, is involved, most likely by indirect means, in the processing and polyadenylation of mtRNA.
Figures

Similar articles
-
Helicase SUV3, polynucleotide phosphorylase, and mitochondrial polyadenylation polymerase form a transient complex to modulate mitochondrial mRNA polyadenylated tail lengths in response to energetic changes.J Biol Chem. 2014 Jun 13;289(24):16727-35. doi: 10.1074/jbc.M113.536540. Epub 2014 Apr 25. J Biol Chem. 2014. PMID: 24770417 Free PMC article.
-
Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poly(A) polymerases and polynucleotide phosphorylase.Plant J. 2009 Jul;59(1):88-99. doi: 10.1111/j.1365-313X.2009.03853.x. Epub 2009 Feb 26. Plant J. 2009. PMID: 19309454
-
Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis.Mol Cell Biol. 2006 Nov;26(22):8475-87. doi: 10.1128/MCB.01002-06. Epub 2006 Sep 11. Mol Cell Biol. 2006. PMID: 16966381 Free PMC article.
-
RNA degradation in human mitochondria: the journey is not finished.Hum Mol Genet. 2024 May 22;33(R1):R26-R33. doi: 10.1093/hmg/ddae043. Hum Mol Genet. 2024. PMID: 38779774 Free PMC article. Review.
-
RNA polyadenylation and decay in mitochondria and chloroplasts.Prog Mol Biol Transl Sci. 2009;85:393-422. doi: 10.1016/S0079-6603(08)00810-6. Prog Mol Biol Transl Sci. 2009. PMID: 19215778 Review.
Cited by
-
Identification of genes potentially regulated by human polynucleotide phosphorylase (hPNPase old-35) using melanoma as a model.PLoS One. 2013 Oct 15;8(10):e76284. doi: 10.1371/journal.pone.0076284. eCollection 2013. PLoS One. 2013. PMID: 24143183 Free PMC article.
-
Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance.Nucleic Acids Res. 2010 Jan;38(1):279-98. doi: 10.1093/nar/gkp903. Epub 2009 Oct 28. Nucleic Acids Res. 2010. PMID: 19864255 Free PMC article.
-
Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition.Nucleic Acids Res. 2010 Jun;38(11):3732-42. doi: 10.1093/nar/gkq068. Epub 2010 Feb 9. Nucleic Acids Res. 2010. PMID: 20144953 Free PMC article.
-
PNPASE and RNA trafficking into mitochondria.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):998-1007. doi: 10.1016/j.bbagrm.2011.10.001. Epub 2011 Oct 13. Biochim Biophys Acta. 2012. PMID: 22023881 Free PMC article. Review.
-
Bacterial/archaeal/organellar polyadenylation.Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):256-76. doi: 10.1002/wrna.51. Wiley Interdiscip Rev RNA. 2011. PMID: 21344039 Free PMC article. Review.
References
-
- Bollenbach, T.J., Schuster, G., Stern, D.B. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. - PubMed
-
- Buttner, K., Wenig, K., Hopfner, K.P. The exosome: A macromolecular cage for controlled RNA degradation. Mol. Microbiol. 2006;61:1372–1379. - PubMed
-
- Chen, H.W., Rainey, R.N., Balatoni, C.E., Dawson, D.W., Troke, J.J., Wasiak, S., Hong, J.S., McBride, H.M., Koehler, C.M., Teitell, M.A., et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
