pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments
- PMID: 18083864
- PMCID: PMC2258600
- DOI: 10.1128/AEM.01194-07
pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments
Erratum in
- Appl Environ Microbiol. 2008 Nov,74(22):7100
Abstract
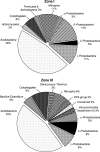
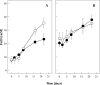
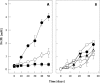
Lakes formed because of coal mining are characterized by low pH and high concentrations of Fe(II) and sulfate. The anoxic sediment is often separated into an upper acidic zone (pH 3; zone I) with large amounts of reactive iron and a deeper slightly acidic zone (pH 5.5; zone III) with smaller amounts of iron. In this study, the impact of pH on the Fe(III)-reducing activities in both of these sediment zones was investigated, and molecular analyses that elucidated the sediment microbial diversity were performed. Fe(II) was formed in zone I and III sediment microcosms at rates that were approximately 710 and 895 nmol cm(-3) day(-1), respectively. A shift to pH 5.3 conditions increased Fe(II) formation in zone I by a factor of 2. A shift to pH 3 conditions inhibited Fe(II) formation in zone III. Clone libraries revealed that the majority of the clones from both zones (approximately 44%) belonged to the Acidobacteria phylum. Since Acidobacterium capsulatum reduced Fe oxides at pHs ranging from 2 to 5, Acidobacteria might be involved in the cycling of iron [corrected]. PCR products specific for species related to Acidiphilium revealed that there were higher numbers of phylotypes related to cultured Acidiphilium or Acidisphaera species in zone III than in zone I. From the PCR products obtained for bioleaching-associated bacteria, only one phylotype with a level of similarity to Acidithiobacillus ferrooxidans of 99% was obtained. Using primer sets specific for Geobacteraceae, PCR products were obtained in higher DNA dilutions from zone III than from zone I. Phylogenetic analysis of clone libraries obtained from Fe(III)-reducing enrichment cultures grown at pH 5.5 revealed that the majority of clones were closely related to members of the Betaproteobacteria, primarily species of Thiomonas. Our results demonstrated that the upper acidic sediment was inhabited by acidophiles or moderate acidophiles which can also reduce Fe(III) under slightly acidic conditions. The majority of Fe(III) reducers inhabiting the slightly acidic sediment had only minor capacities to be active under acidic conditions.
Figures




References
-
- Achouak, W., P. Normand, and T. Heulin. 1999. Comparative phylogeny of rrs and nifH genes in the Bacillaceae. Int. J. Syst. Bacteriol. 49:961-967. - PubMed
-
- Akob, D. M., H. J. Mills, and J. E. Kostka. 2007. Metabolically active microbial communities in uranium contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95-107. - PubMed
-
- Alef, K. 1991. Methodenhandbuch Bodenmikrobiologie: Aktivitäten, Biomasse, Differenzierung. Ecomed Verlagsgesellschaft mgH, Landsberg/Lech, Germany.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and J. D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

