N-glycan modification in Aspergillus species
- PMID: 18083888
- PMCID: PMC2258586
- DOI: 10.1128/AEM.01058-07
N-glycan modification in Aspergillus species
Abstract
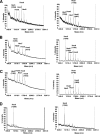
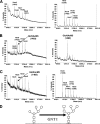
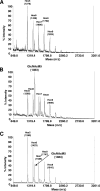
The production by filamentous fungi of therapeutic glycoproteins intended for use in mammals is held back by the inherent difference in protein N-glycosylation and by the inability of the fungal cell to modify proteins with mammalian glycosylation structures. Here, we report protein N-glycan engineering in two Aspergillus species. We functionally expressed in the fungal hosts heterologous chimeric fusion proteins containing different localization peptides and catalytic domains. This strategy allowed the isolation of a strain with a functional alpha-1,2-mannosidase producing increased amounts of N-glycans of the Man5GlcNAc2 type. This strain was further engineered by the introduction of a functional GlcNAc transferase I construct yielding GlcNAcMan5GlcNac2 N-glycans. Additionally, we deleted algC genes coding for an enzyme involved in an early step of the fungal glycosylation pathway yielding Man3GlcNAc2 N-glycans. This modification of fungal glycosylation is a step toward the ability to produce humanized complex N-glycans on therapeutic proteins in filamentous fungi.
Figures



Similar articles
-
Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris.Proc Natl Acad Sci U S A. 2003 Apr 29;100(9):5022-7. doi: 10.1073/pnas.0931263100. Epub 2003 Apr 17. Proc Natl Acad Sci U S A. 2003. PMID: 12702754 Free PMC article.
-
Glycoengineering of Aspergillus nidulans to produce precursors for humanized N-glycan structures.Metab Eng. 2021 Sep;67:153-163. doi: 10.1016/j.ymben.2021.06.001. Epub 2021 Jun 24. Metab Eng. 2021. PMID: 34174425
-
Engineering the protein N-glycosylation pathway in insect cells for production of biantennary, complex N-glycans.Biochemistry. 2002 Dec 17;41(50):15093-104. doi: 10.1021/bi026455d. Biochemistry. 2002. PMID: 12475259 Free PMC article.
-
Engineering Yarrowia lipolytica to produce glycoproteins homogeneously modified with the universal Man3GlcNAc2 N-glycan core.PLoS One. 2012;7(6):e39976. doi: 10.1371/journal.pone.0039976. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768188 Free PMC article.
-
Off-target glycans encountered along the synthetic biology route toward humanized N-glycans in Pichia pastoris.Biotechnol Bioeng. 2020 Aug;117(8):2479-2488. doi: 10.1002/bit.27375. Epub 2020 May 26. Biotechnol Bioeng. 2020. PMID: 32374435
Cited by
-
Homologue expression of a β-xylosidase from native Aspergillus niger.J Ind Microbiol Biotechnol. 2011 Sep;38(9):1311-9. doi: 10.1007/s10295-010-0912-5. Epub 2010 Nov 30. J Ind Microbiol Biotechnol. 2011. PMID: 21116681
-
The Impact of Glycoengineering on the Endoplasmic Reticulum Quality Control System in Yeasts.Front Mol Biosci. 2022 Jun 2;9:910709. doi: 10.3389/fmolb.2022.910709. eCollection 2022. Front Mol Biosci. 2022. PMID: 35720120 Free PMC article. Review.
-
Unlocking the magic in mycelium: Using synthetic biology to optimize filamentous fungi for biomanufacturing and sustainability.Mater Today Bio. 2023 Jan 21;19:100560. doi: 10.1016/j.mtbio.2023.100560. eCollection 2023 Apr. Mater Today Bio. 2023. PMID: 36756210 Free PMC article.
-
Engineering of Yeast Glycoprotein Expression.Adv Biochem Eng Biotechnol. 2021;175:93-135. doi: 10.1007/10_2018_69. Adv Biochem Eng Biotechnol. 2021. PMID: 30397726
-
Engineering the supply chain for protein production/secretion in yeasts and mammalian cells.J Ind Microbiol Biotechnol. 2015 Mar;42(3):453-64. doi: 10.1007/s10295-014-1569-2. Epub 2015 Jan 6. J Ind Microbiol Biotechnol. 2015. PMID: 25561318 Review.
References
-
- Aleshin, A. E., L. M. Firsov, and R. B. Honzatko. 1994. Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4- Å resolution. J. Biol. Chem. 269:15631-9. - PubMed
-
- Archer, D. B., and J. F. Peberdy. 1997. The molecular biology of secreted enzyme production by fungi. Crit. Rev. Biotechnol. 17:273-306. - PubMed
-
- Aunstrup, K. 1977. Production of industrial enzymes, p. 151-171. In J. Meyrath and J. D. Bu'Lock (ed.), Biotechnology and fungal differentiation. FEMS Symposium vol. 4. Academic Press, New York, NY.
-
- Bobrowicz, P., R. C. Davidson, H. Li, T. I. Potgieter, J. H. Nett, S. R. Hamilton, T. A. Stadheim, R. G. Miele, B. Bobrowicz, T. Mitchell, S. Rausch, E. Renfer, and S. Wildt. 2004. Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: production of complex humanized glycoproteins with terminal galactose. Glycobiology 14:757-766. - PubMed
-
- Callewaert, N., W. Laroy, H. Cadirgi, S. Geysens, X. Saelens, W. Min Jou, and R. Contreras. 2001. Use of HDEL-tagged Trichoderma reesei mannosyl oligosaccharide 1,2-alpha-d-mannosidase for N-glycan engineering in Pichia pastoris. FEBS Lett. 503:173-178. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

