Solution structures of 2 : 1 and 1 : 1 DNA polymerase-DNA complexes probed by ultracentrifugation and small-angle X-ray scattering
- PMID: 18084022
- PMCID: PMC2241917
- DOI: 10.1093/nar/gkm1101
Solution structures of 2 : 1 and 1 : 1 DNA polymerase-DNA complexes probed by ultracentrifugation and small-angle X-ray scattering
Abstract
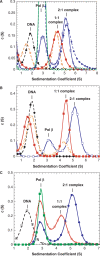
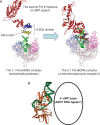
We report small-angle X-ray scattering (SAXS) and sedimentation velocity (SV) studies on the enzyme-DNA complexes of rat DNA polymerase beta (Pol beta) and African swine fever virus DNA polymerase X (ASFV Pol X) with one-nucleotide gapped DNA. The results indicated formation of a 2 : 1 Pol beta-DNA complex, whereas only 1 : 1 Pol X-DNA complex was observed. Three-dimensional structural models for the 2 : 1 Pol beta-DNA and 1 : 1 Pol X-DNA complexes were generated from the SAXS experimental data to correlate with the functions of the DNA polymerases. The former indicates interactions of the 8 kDa 5'-dRP lyase domain of the second Pol beta molecule with the active site of the 1 : 1 Pol beta-DNA complex, while the latter demonstrates how ASFV Pol X binds DNA in the absence of DNA-binding motif(s). As ASFV Pol X has no 5'-dRP lyase domain, it is reasonable not to form a 2 : 1 complex. Based on the enhanced activities of the 2 : 1 complex and the observation that the 8 kDa domain is not in an optimal configuration for the 5'-dRP lyase reaction in the crystal structures of the closed ternary enzyme-DNA-dNTP complexes, we propose that the asymmetric 2 : 1 Pol beta-DNA complex enhances the function of Pol beta.
Figures





References
-
- Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. - PubMed
-
- Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. - PubMed
-
- Rajendran S, Jezewska MJ, Bujalowski W. Recognition of template-primer and gapped DNA substrates by the human DNA polymerase beta. J. Mol. Biol. 2001;308:477–500. - PubMed
-
- Bailey MF, Van der Schans EJ, Millar DP. Dimerization of the Klenow fragment of Escherichia coli DNA polymerase I is linked to its mode of DNA binding. Biochemistry. 2007;46:8085–8099. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

