Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells
- PMID: 18084025
- PMCID: PMC2241897
- DOI: 10.1093/nar/gkm1105
Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells
Abstract
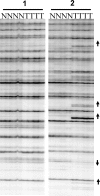
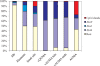
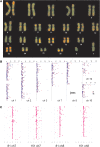
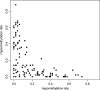
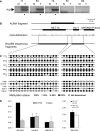
Methylation of the cytosine is the most frequent epigenetic modification of DNA in mammalian cells. In humans, most of the methylated cytosines are found in CpG-rich sequences within tandem and interspersed repeats that make up to 45% of the human genome, being Alu repeats the most common family. Demethylation of Alu elements occurs in aging and cancer processes and has been associated with gene reactivation and genomic instability. By targeting the unmethylated SmaI site within the Alu sequence as a surrogate marker, we have quantified and identified unmethylated Alu elements on the genomic scale. Normal colon epithelial cells contain in average 25 486 +/- 10 157 unmethylated Alu's per haploid genome, while in tumor cells this figure is 41 995 +/- 17 187 (P = 0.004). There is an inverse relationship in Alu families with respect to their age and methylation status: the youngest elements exhibit the highest prevalence of the SmaI site (AluY: 42%; AluS: 18%, AluJ: 5%) but the lower rates of unmethylation (AluY: 1.65%; AluS: 3.1%, AluJ: 12%). Data are consistent with a stronger silencing pressure on the youngest repetitive elements, which are closer to genes. Further insights into the functional implications of atypical unmethylation states in Alu elements will surely contribute to decipher genomic organization and gene regulation in complex organisms.
Figures

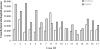
References
-
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. - PubMed
-
- Zuckerkandl E. Why so many noncoding nucleotides? The eukaryote genome as an epigenetic machine. Genetica. 2002;115:105–129. - PubMed
-
- Kreahling J, Graveley BR. The origins and implications of Aluternative splicing. Trends Genet. 2004;20:1–4. - PubMed
-
- Jasinska A, Krzyzosiak WJ. Repetitive sequences that shape the human transcriptome. FEBS Lett. 2004;567:136–141. - PubMed
-
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

