Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor
- PMID: 18084030
- PMCID: PMC2241913
- DOI: 10.1093/nar/gkm1096
Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor
Abstract
The ability to sense and respond to environmental and physiological signals is critical for the survival of the soil-dwelling Gram-positive bacterium Streptomyces coelicolor. Nutrient deprivation triggers the onset of a complex morphological differentiation process that involves the raising of aerial hyphae and formation of spore chains, and coincides with the production of a diverse array of clinically relevant antibiotics and other secondary metabolites. These processes are tightly regulated; however, the genes and signals involved have not been fully elucidated. Here, we report a novel tRNA cleavage event that follows the same temporal regulation as morphological and physiological differentiation, and is growth medium dependent. All tRNAs appear to be susceptible to cleavage; however, there appears to be a bias towards increased cleavage of those tRNAs that specify highly utilized codons. In contrast to what has been observed in eukaryotes, accumulation of tRNA halves in S. coelicolor is not significantly affected by amino acid starvation, and is also not affected by induction of the stringent response or inhibition of ribosome function. Mutants defective in aerial development and antibiotic production exhibit altered tRNA cleavage profiles relative to wild-type strains.
Figures
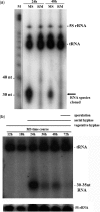

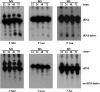


References
-
- Elliot MA, Buttner MJ, Nodwell JR. Multicellular development in Streptomyces. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. Washington, D.C.: ASM Press; 2007. pp. 419–438.
-
- Claessen D, de Jong W, Dijkhuizen L, Wosten HAB. Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 2006;14:313–319. - PubMed
-
- Kelemen GH, Buttner MJ. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1998;1:656–662. - PubMed
-
- Pope MK, Green BD, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 1996;19:747–756. - PubMed
-
- Rigali S, Nothaft H, Noens EEE, Schlicht M, Colson S, Müller M, Joris B, Koerten HK, Hopwood DA, et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 2006;61:1237–1251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

