Co-circulation and evolution of polioviruses and species C enteroviruses in a district of Madagascar
- PMID: 18085822
- PMCID: PMC2134956
- DOI: 10.1371/journal.ppat.0030191
Co-circulation and evolution of polioviruses and species C enteroviruses in a district of Madagascar
Abstract
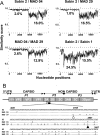
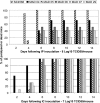
Between October 2001 and April 2002, five cases of acute flaccid paralysis (AFP) associated with type 2 vaccine-derived polioviruses (VDPVs) were reported in the southern province of the Republic of Madagascar. To determine viral factors that favor the emergence of these pathogenic VDPVs, we analyzed in detail their genomic and phenotypic characteristics and compared them with co-circulating enteroviruses. These VDPVs appeared to belong to two independent recombinant lineages with sequences from the type 2 strain of the oral poliovaccine (OPV) in the 5'-half of the genome and sequences derived from unidentified species C enteroviruses (HEV-C) in the 3'-half. VDPV strains showed characteristics similar to those of wild neurovirulent viruses including neurovirulence in poliovirus-receptor transgenic mice. We looked for other VDPVs and for circulating enteroviruses in 316 stools collected from healthy children living in the small area where most of the AFP cases occurred. We found vaccine PVs, two VDPVs similar to those found in AFP cases, some echoviruses, and above all, many serotypes of coxsackie A viruses belonging to HEV-C, with substantial genetic diversity. Several coxsackie viruses A17 and A13 carried nucleotide sequences closely related to the 2C and the 3D(pol) coding regions of the VDPVs, respectively. There was also evidence of multiple genetic recombination events among the HEV-C resulting in numerous recombinant genotypes. This indicates that co-circulation of HEV-C and OPV strains is associated with evolution by recombination, resulting in unexpectedly extensive viral diversity in small human populations in some tropical regions. This probably contributed to the emergence of recombinant VDPVs. These findings give further insight into viral ecosystems and the evolutionary processes that shape viral biodiversity.
Conflict of interest statement
Figures


References
-
- Pallansch M, Roos R. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe D, Howley P, editors. Fields virology. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 723–775.
-
- Anonymous. Progress toward interruption of wild poliovirus transmission worldwide, January 2006–May 2007. Morb Mortal Wkly Rep. 2007;56:682–685. - PubMed
-
- Minor PD. The molecular biology of poliovaccines. J Gen Virol. 1992;73:3065–3077. - PubMed
-
- Dahourou G, Guillot S, Le Gall O, Crainic R. Genetic recombination in wild-type poliovirus. J Gen Virol. 2002;83:3103–3110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

