Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families
- PMID: 18086378
- PMCID: PMC2705969
- DOI: 10.1016/j.imbio.2007.09.018
Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families
Abstract
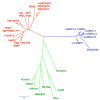
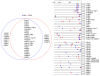
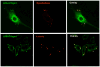
Vertebrates have evolved complex immune specificity repertoires beyond the primordial components found in lower multi-cellular organisms to combat microbial infections. The type II interferon (IFN-gamma) pathway represents one such system, bridging innate and acquired immunity and providing host protection in a cell-autonomous manner. Recent large-scale transcriptome analyses of IFN-gamma-dependent gene expression in effector cells such as macrophages have highlighted the prominence of two families of GTPases -- p47 IRGs and p65 GBPs -- that are now beginning to emerge as major determinants of antimicrobial resistance. Here we discuss the recent clarification of known family members, their cellular biochemistry and host defense functions as a means to understanding the complex innate immune response engendered in higher vertebrates such as humans and mice.
Figures


References
-
- Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256:8–14. - PubMed
-
- Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, Futterer A, Pfeffer K, Howard JC. Two families of GTPases dominate the complex cellular response to IFN-gamma. J Immunol. 1998;161:6715–6723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

