Tetrameric restriction enzymes: expansion to the GIY-YIG nuclease family
- PMID: 18086711
- PMCID: PMC2241918
- DOI: 10.1093/nar/gkm1090
Tetrameric restriction enzymes: expansion to the GIY-YIG nuclease family
Abstract
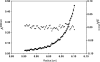
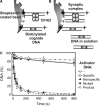
The GIY-YIG nuclease domain was originally identified in homing endonucleases and enzymes involved in DNA repair and recombination. Many of the GIY-YIG family enzymes are functional as monomers. We show here that the Cfr42I restriction endonuclease which belongs to the GIY-YIG family and recognizes the symmetric sequence 5'-CCGC/GG-3' ('/' indicates the cleavage site) is a tetramer in solution. Moreover, biochemical and kinetic studies provided here demonstrate that the Cfr42I tetramer is catalytically active only upon simultaneous binding of two copies of its recognition sequence. In that respect Cfr42I resembles the homotetrameric Type IIF restriction enzymes that belong to the distinct PD-(E/D)XK nuclease superfamily. Unlike the PD-(E/D)XK enzymes, the GIY-YIG nuclease Cfr42I accommodates an extremely wide selection of metal-ion cofactors, including Mg2+, Mn2+, Co2+, Zn2+, Ni2+, Cu2+ and Ca2+. To our knowledge, Cfr42I is the first tetrameric GIY-YIG family enzyme. Similar structural arrangement and phenotypes displayed by restriction enzymes of the PD-(E/D)XK and GIY-YIG nuclease families point to the functional significance of tetramerization.
Figures
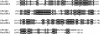




Similar articles
-
Oligomeric structure diversity within the GIY-YIG nuclease family.J Mol Biol. 2009 Mar 20;387(1):10-6. doi: 10.1016/j.jmb.2009.01.048. Epub 2009 Jan 30. J Mol Biol. 2009. PMID: 19361436
-
Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily.BMC Struct Biol. 2007 Jul 12;7:48. doi: 10.1186/1472-6807-7-48. BMC Struct Biol. 2007. PMID: 17626614 Free PMC article.
-
Type II restriction endonuclease R.Hpy188I belongs to the GIY-YIG nuclease superfamily, but exhibits an unusual active site.BMC Struct Biol. 2008 Nov 14;8:48. doi: 10.1186/1472-6807-8-48. BMC Struct Biol. 2008. PMID: 19014591 Free PMC article.
-
Catalytic mechanisms of restriction and homing endonucleases.Biochemistry. 2002 Nov 26;41(47):13851-60. doi: 10.1021/bi020467h. Biochemistry. 2002. PMID: 12437341 Review.
-
Type II restriction endonucleases: structure and mechanism.Cell Mol Life Sci. 2005 Mar;62(6):685-707. doi: 10.1007/s00018-004-4513-1. Cell Mol Life Sci. 2005. PMID: 15770420 Free PMC article. Review.
Cited by
-
CgII cleaves DNA using a mechanism distinct from other ATP-dependent restriction endonucleases.Nucleic Acids Res. 2017 Aug 21;45(14):8435-8447. doi: 10.1093/nar/gkx580. Nucleic Acids Res. 2017. PMID: 28854738 Free PMC article.
-
A unified genetic, computational and experimental framework identifies functionally relevant residues of the homing endonuclease I-BmoI.Nucleic Acids Res. 2010 Apr;38(7):2411-27. doi: 10.1093/nar/gkp1223. Epub 2010 Jan 8. Nucleic Acids Res. 2010. PMID: 20061372 Free PMC article.
-
Bacteriophage T4 endonuclease II, a promiscuous GIY-YIG nuclease, binds as a tetramer to two DNA substrates.Nucleic Acids Res. 2009 Oct;37(18):6174-83. doi: 10.1093/nar/gkp652. Epub 2009 Aug 7. Nucleic Acids Res. 2009. PMID: 19666720 Free PMC article.
-
Folding, DNA recognition, and function of GIY-YIG endonucleases: crystal structures of R.Eco29kI.Structure. 2010 Oct 13;18(10):1321-31. doi: 10.1016/j.str.2010.07.006. Epub 2010 Aug 26. Structure. 2010. PMID: 20800503 Free PMC article.
-
Amino acid residues in the GIY-YIG endonuclease II of phage T4 affecting sequence recognition and binding as well as catalysis.J Bacteriol. 2008 Aug;190(16):5533-44. doi: 10.1128/JB.00094-08. Epub 2008 Jun 6. J Bacteriol. 2008. PMID: 18539732 Free PMC article.
References
-
- Sapranauskas R, Sasnauskas G, Lagunavicius A, Vilkaitis G, Lubys A, Siksnys V. Novel subtype of type IIs restriction enzymes. BfiI endonuclease exhibits similarities to the EDTA-resistant nuclease Nuc of Salmonella typhimurium. J. Biol. Chem. 2000;275:30878–30885. - PubMed
-
- Bujnicki JM, Radlinska M, Rychlewski L. Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem. Sci. 2001;26:9–11. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

