Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II
- PMID: 18086892
- PMCID: PMC2258746
- DOI: 10.1128/MCB.01733-07
Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II
Abstract
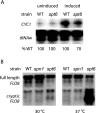
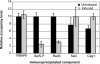
We investigated the timing of the recruitment of Spn1 and its partner, Spt6, to the CYC1 gene. Like TATA binding protein and RNA polymerase II (RNAPII), Spn1 is constitutively recruited to the CYC1 promoter, although levels of transcription from this gene, which is regulated postrecruitment of RNAPII, are low. In contrast, Spt6 appears only after growth in conditions in which the gene is highly transcribed. Spn1 recruitment is via interaction with RNAPII, since an spn1 mutant defective for interaction with RNAPII is not targeted to the promoter, and Spn1 is necessary for Spt6 recruitment. Through a targeted genetic screen, strong and specific antagonizing interactions between SPN1 and genes encoding Swi/Snf subunits were identified. Like Spt6, Swi/Snf appears at CYC1 only after activation of the gene. However, Spt6 significantly precedes Swi/Snf occupancy at the promoter. In the absence of Spn1 recruitment, Swi/Snf is constitutively found at the promoter. These observations support a model whereby Spn1 negatively regulates RNAPII transcriptional activity by inhibiting recruitment of Swi/Snf to the CYC1 promoter, and this inhibition is abrogated by the Spn1-Spt6 interaction. These findings link Spn1 functions to the transition from an inactive to an actively transcribing RNAPII complex at a postrecruitment-regulated promoter.
Figures







Similar articles
-
Spt6-Spn1 interaction is required for RNA polymerase II association and precise nucleosome positioning along transcribed genes.J Biol Chem. 2025 May;301(5):108436. doi: 10.1016/j.jbc.2025.108436. Epub 2025 Mar 22. J Biol Chem. 2025. PMID: 40127868 Free PMC article.
-
The elongation factor Spn1 is a multi-functional chromatin binding protein.Nucleic Acids Res. 2018 Mar 16;46(5):2321-2334. doi: 10.1093/nar/gkx1305. Nucleic Acids Res. 2018. PMID: 29300974 Free PMC article.
-
Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast.J Biol Chem. 2014 May 23;289(21):14981-95. doi: 10.1074/jbc.M113.529354. Epub 2014 Apr 11. J Biol Chem. 2014. PMID: 24727477 Free PMC article.
-
Insights into Spt6: a histone chaperone that functions in transcription, DNA replication, and genome stability.Trends Genet. 2023 Nov;39(11):858-872. doi: 10.1016/j.tig.2023.06.008. Epub 2023 Jul 20. Trends Genet. 2023. PMID: 37481442 Free PMC article. Review.
-
Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation.Biochim Biophys Acta. 2011 Jan;1809(1):34-45. doi: 10.1016/j.bbagrm.2010.11.001. Epub 2010 Nov 13. Biochim Biophys Acta. 2011. PMID: 21081187 Free PMC article. Review.
Cited by
-
Genome Instability Is Promoted by the Chromatin-Binding Protein Spn1 in Saccharomyces cerevisiae.Genetics. 2018 Dec;210(4):1227-1237. doi: 10.1534/genetics.118.301600. Epub 2018 Oct 9. Genetics. 2018. PMID: 30301740 Free PMC article.
-
The head module of Mediator directs activation of preloaded RNAPII in vivo.Nucleic Acids Res. 2013 Dec;41(22):10124-34. doi: 10.1093/nar/gkt796. Epub 2013 Sep 4. Nucleic Acids Res. 2013. PMID: 24005039 Free PMC article.
-
The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions.Mol Cell. 2010 Mar 26;37(6):834-42. doi: 10.1016/j.molcel.2010.01.037. Mol Cell. 2010. PMID: 20347425 Free PMC article.
-
Emerging Views on the CTD Code.Genet Res Int. 2012;2012:347214. doi: 10.1155/2012/347214. Epub 2012 Feb 26. Genet Res Int. 2012. PMID: 22567385 Free PMC article.
-
The Abundant Histone Chaperones Spt6 and FACT Collaborate to Assemble, Inspect, and Maintain Chromatin Structure in Saccharomyces cerevisiae.Genetics. 2015 Nov;201(3):1031-45. doi: 10.1534/genetics.115.180794. Epub 2015 Sep 28. Genetics. 2015. PMID: 26416482 Free PMC article.
References
-
- Adelman, K., M. T. Marr, J. Werner, A. Saunders, Z. Ni, E. D. Andrulis, and J. T. Lis. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17103-112. - PubMed
-
- Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21405-416. - PubMed
-
- Awrey, D. E., N. Shimasaki, C. Koth, R. Weilbaecher, V. Olmsted, S. Kazanis, X. Shan, J. Arellano, C. H. Arrowsmith, C. M. Kane, and A. M. Edwards. 1998. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J. Biol. Chem. 27322595-22605. - PubMed
-
- Awrey, D. E., R. G. Weilbaecher, S. A. Hemming, S. M. Orlicky, C. M. Kane, and A. M. Edwards. 1997. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 27214747-14754. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
