Gene and genon concept: coding versus regulation. A conceptual and information-theoretic analysis of genetic storage and expression in the light of modern molecular biology
- PMID: 18087760
- PMCID: PMC2242853
- DOI: 10.1007/s12064-007-0012-x
Gene and genon concept: coding versus regulation. A conceptual and information-theoretic analysis of genetic storage and expression in the light of modern molecular biology
Abstract
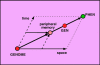
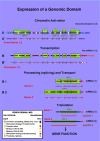
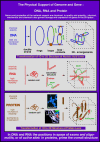
We analyse here the definition of the gene in order to distinguish, on the basis of modern insight in molecular biology, what the gene is coding for, namely a specific polypeptide, and how its expression is realized and controlled. Before the coding role of the DNA was discovered, a gene was identified with a specific phenotypic trait, from Mendel through Morgan up to Benzer. Subsequently, however, molecular biologists ventured to define a gene at the level of the DNA sequence in terms of coding. As is becoming ever more evident, the relations between information stored at DNA level and functional products are very intricate, and the regulatory aspects are as important and essential as the information coding for products. This approach led, thus, to a conceptual hybrid that confused coding, regulation and functional aspects. In this essay, we develop a definition of the gene that once again starts from the functional aspect. A cellular function can be represented by a polypeptide or an RNA. In the case of the polypeptide, its biochemical identity is determined by the mRNA prior to translation, and that is where we locate the gene. The steps from specific, but possibly separated sequence fragments at DNA level to that final mRNA then can be analysed in terms of regulation. For that purpose, we coin the new term "genon". In that manner, we can clearly separate product and regulative information while keeping the fundamental relation between coding and function without the need to introduce a conceptual hybrid. In mRNA, the program regulating the expression of a gene is superimposed onto and added to the coding sequence in cis - we call it the genon. The complementary external control of a given mRNA by trans-acting factors is incorporated in its transgenon. A consequence of this definition is that, in eukaryotes, the gene is, in most cases, not yet present at DNA level. Rather, it is assembled by RNA processing, including differential splicing, from various pieces, as steered by the genon. It emerges finally as an uninterrupted nucleic acid sequence at mRNA level just prior to translation, in faithful correspondence with the amino acid sequence to be produced as a polypeptide. After translation, the genon has fulfilled its role and expires. The distinction between the protein coding information as materialised in the final polypeptide and the processing information represented by the genon allows us to set up a new information theoretic scheme. The standard sequence information determined by the genetic code expresses the relation between coding sequence and product. Backward analysis asks from which coding region in the DNA a given polypeptide originates. The (more interesting) forward analysis asks in how many polypeptides of how many different types a given DNA segment is expressed. This concerns the control of the expression process for which we have introduced the genon concept. Thus, the information theoretic analysis can capture the complementary aspects of coding and regulation, of gene and genon.
Figures






Comment in
-
Commentary on Scherrer and Jost (2007) Gene and genon concept: coding versus regulation.Theory Biosci. 2009 Aug;128(3):153-4; discussion 171-7. doi: 10.1007/s12064-009-0073-0. Epub 2009 Aug 21. Theory Biosci. 2009. PMID: 19697074 No abstract available.
-
Comments on the paper by K. Scherrer and J. Jost "Gene and genon" concept: coding versus regulation.Theory Biosci. 2009 Aug;128(3):155-6; discussion 171-7. doi: 10.1007/s12064-009-0070-3. Theory Biosci. 2009. PMID: 19730918 No abstract available.
-
Comments on "Gene and genon concept" by K. Scherrer and J. Jost.Theory Biosci. 2009 Aug;128(3):163-4; discussion 171-7. doi: 10.1007/s12064-009-0068-x. Theory Biosci. 2009. PMID: 19779755 No abstract available.
Similar articles
-
Commentary on Scherrer and Jost (2007) Gene and genon concept: coding versus regulation.Theory Biosci. 2009 Aug;128(3):153-4; discussion 171-7. doi: 10.1007/s12064-009-0073-0. Epub 2009 Aug 21. Theory Biosci. 2009. PMID: 19697074 No abstract available.
-
Comments on the paper by K. Scherrer and J. Jost "Gene and genon" concept: coding versus regulation.Theory Biosci. 2009 Aug;128(3):155-6; discussion 171-7. doi: 10.1007/s12064-009-0070-3. Theory Biosci. 2009. PMID: 19730918 No abstract available.
-
The gene and the genon concept: a functional and information-theoretic analysis.Mol Syst Biol. 2007;3:87. doi: 10.1038/msb4100123. Epub 2007 Mar 13. Mol Syst Biol. 2007. PMID: 17353929 Free PMC article. Review.
-
Primary transcripts: From the discovery of RNA processing to current concepts of gene expression - Review.Exp Cell Res. 2018 Dec 15;373(1-2):1-33. doi: 10.1016/j.yexcr.2018.09.011. Epub 2018 Sep 26. Exp Cell Res. 2018. PMID: 30266658 Review.
-
Comments on "Gene and genon concept" by K. Scherrer and J. Jost.Theory Biosci. 2009 Aug;128(3):163-4; discussion 171-7. doi: 10.1007/s12064-009-0068-x. Theory Biosci. 2009. PMID: 19779755 No abstract available.
Cited by
-
Biology, geometry and information.Theory Biosci. 2022 Jun;141(2):65-71. doi: 10.1007/s12064-021-00351-9. Epub 2021 Jun 11. Theory Biosci. 2022. PMID: 34117616 Free PMC article.
-
Defining genes: a computational framework.Theory Biosci. 2009 Aug;128(3):165-70. doi: 10.1007/s12064-009-0067-y. Epub 2009 Jun 26. Theory Biosci. 2009. PMID: 19557452 Free PMC article.
-
Commentary on Scherrer and Jost (2007) Gene and genon concept: coding versus regulation.Theory Biosci. 2009 Aug;128(3):153-4; discussion 171-7. doi: 10.1007/s12064-009-0073-0. Epub 2009 Aug 21. Theory Biosci. 2009. PMID: 19697074 No abstract available.
-
The Evolving Definition of the Term "Gene".Genetics. 2017 Apr;205(4):1353-1364. doi: 10.1534/genetics.116.196956. Genetics. 2017. PMID: 28360126 Free PMC article.
-
Comments on the paper by K. Scherrer and J. Jost "Gene and genon" concept: coding versus regulation.Theory Biosci. 2009 Aug;128(3):155-6; discussion 171-7. doi: 10.1007/s12064-009-0070-3. Theory Biosci. 2009. PMID: 19730918 No abstract available.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '17213034', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17213034/'}]}
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J et al (2006) Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem 50:223–272. Review - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '6790245', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/6790245/'}]}
- Ananiev E, Barsky V, Ilyin Y, Churikov N (1981) Localization of nucleoli in Drosophila melanogaster polytene chromosomes. Chromosoma 81:619–628 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PMC', 'value': 'PMC59777', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC59777/'}, {'type': 'PubMed', 'value': '11593024', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11593024/'}]}
- Anguita E, Johnson CA, Wood WG, Turner BM, Higgs DR (2001) Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc Natl Acad Sci USA 98:12114–12119 - PMC - PubMed
-
- None
- Ashby WR (1956) An introduction to cybernetics. Chapman and Hall, London
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '9216087', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9216087/'}]}
- Arcangeletti C, Sütterlin R, Aebi U, De Conto F, Missorini S, Chezzi C, Scherrer K (1997) Visualization of prosomes (MCP-proteasomes), intermediate filament and actin networks by “instantaneous fixation” preserving the cytoskeleton. J Struct Biol 119:35–58 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

