Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data
- PMID: 18088415
- PMCID: PMC2225423
- DOI: 10.1186/1471-2407-7-226
Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data
Abstract
Background: Chromosomal abnormalities have been associated with most human malignancies, with gains and losses on some genomic regions associated with particular entities.
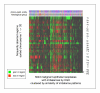
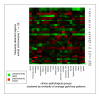
Methods: Of the 15429 cases collected for the Progenetix molecular-cytogenetic database, 5918 malignant epithelial neoplasias analyzed by chromosomal Comparative Genomic Hybridization (CGH) were selected for further evaluation. For the 22 clinico-pathological entities with more than 50 cases, summary profiles for genomic imbalances were generated from case specific data and analyzed.
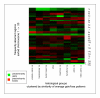
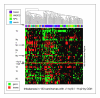
Results: With large variation in overall genomic instability, recurring genomic gains and losses were prominent. Most entities showed frequent gains involving 8q2, while gains on 20q, 1q, 3q, 5p, 7q and 17q were frequent in different entities. Loss "hot spots" included 3p, 4q, 13q, 17p and 18q among others. Related average imbalance patterns were found for clinically distinct entities, e.g. hepatocellular carcinomas (ca.) and ductal breast ca., as well as for histologically related entities (squamous cell ca. of different sites).
Conclusion: Although considerable case-by-case variation of genomic profiles can be found by CGH in epithelial malignancies, a limited set of variously combined chromosomal imbalances may be typical for carcinogenesis. Focus on the respective regions should aid in target gene detection and pathway deduction.
Figures
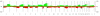
References
-
- Crossen PE. Giemsa banding patterns of human chromosomes. Clin Genet. 1972;3:169–179. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

