Mosquito transcriptome changes and filarial worm resistance in Armigeres subalbatus
- PMID: 18088420
- PMCID: PMC2234435
- DOI: 10.1186/1471-2164-8-463
Mosquito transcriptome changes and filarial worm resistance in Armigeres subalbatus
Abstract
Background: Armigeres subalbatus is a natural vector of the filarial worm Brugia pahangi, but it rapidly and proficiently kills Brugia malayi microfilariae by melanotic encapsulation. Because B. malayi and B. pahangi are morphologically and biologically similar, the Armigeres-Brugia system serves as a valuable model for studying the resistance mechanisms in mosquito vectors. We have initiated transcriptome profiling studies in Ar. subalbatus to identify molecular components involved in B. malayi refractoriness.
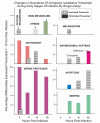
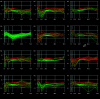
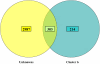
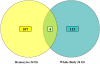
Results: These initial studies assessed the transcriptional response of Ar. subalbatus to B. malayi at 1, 3, 6, 12, 24, 48, and 72 hrs after an infective blood feed. In this investigation, we initiated the first holistic study conducted on the anti-filarial worm immune response in order to effectively explore the functional roles of immune-response genes following a natural exposure to the parasite. Studies assessing the transcriptional response revealed the involvement of unknown and conserved unknowns, cytoskeletal and structural components, and stress and immune responsive factors. The data show that the anti-filarial worm immune response by Ar. subalbatus to be a highly complex, tissue-specific process involving varied effector responses working in concert with blood cell-mediated melanization.
Conclusion: This initial study provides a foundation and direction for future studies, which will more fully dissect the nature of the anti-filarial worm immune response in this mosquito-parasite system. The study also argues for continued studies with RNA generated from both hemocytes and whole bodies to fully expound the nature of the anti-filarial worm immune response.
Figures

Similar articles
-
Mosquito transcriptome profiles and filarial worm susceptibility in Armigeres subalbatus.PLoS Negl Trop Dis. 2010 Apr 20;4(4):e666. doi: 10.1371/journal.pntd.0000666. PLoS Negl Trop Dis. 2010. PMID: 20421927 Free PMC article.
-
Response of Armigeres subalbatus (Diptera: Culicidae) to intraperitoneally isolated Brugia spp. microfilariae.J Med Entomol. 2007 Mar;44(2):295-8. doi: 10.1603/0022-2585(2007)44[295:roasdc]2.0.co;2. J Med Entomol. 2007. PMID: 17427699
-
Brugia malayi and Brugia pahangi: inherent difference in immune activation in the mosquitoes Armigeres subalbatus and Aedes aegypti.J Parasitol. 1989 Feb;75(1):76-81. J Parasitol. 1989. PMID: 2563767
-
Immunity to eukaryotic parasites in vector insects.Curr Opin Immunol. 1996 Feb;8(1):14-9. doi: 10.1016/s0952-7915(96)80099-9. Curr Opin Immunol. 1996. PMID: 8729441 Review.
-
Infection barriers and responses in mosquito-filarial worm interactions.Curr Opin Insect Sci. 2014 Sep;3:37-42. doi: 10.1016/j.cois.2014.08.006. Epub 2014 Aug 15. Curr Opin Insect Sci. 2014. PMID: 32846673 Review.
Cited by
-
The roles of serpins in mosquito immunology and physiology.J Insect Physiol. 2013 Feb;59(2):138-47. doi: 10.1016/j.jinsphys.2012.08.015. Epub 2012 Sep 5. J Insect Physiol. 2013. PMID: 22960307 Free PMC article. Review.
-
Molecular characterization of immune responses of Helicoverpa armigera to infection with the mermithid nematode Ovomermis sinensis.BMC Genomics. 2019 Feb 27;20(1):161. doi: 10.1186/s12864-019-5544-1. BMC Genomics. 2019. PMID: 30813894 Free PMC article.
-
Evolution of pathogen tolerance and emerging infections: A missing experimental paradigm.Elife. 2021 Sep 21;10:e68874. doi: 10.7554/eLife.68874. Elife. 2021. PMID: 34544548 Free PMC article. Review.
-
Mosquito infection responses to developing filarial worms.PLoS Negl Trop Dis. 2009 Oct 13;3(10):e529. doi: 10.1371/journal.pntd.0000529. PLoS Negl Trop Dis. 2009. PMID: 19823571 Free PMC article.
-
C-Type Lectin-20 Interacts with ALP1 Receptor to Reduce Cry Toxicity in Aedes aegypti.Toxins (Basel). 2018 Sep 25;10(10):390. doi: 10.3390/toxins10100390. Toxins (Basel). 2018. PMID: 30257487 Free PMC article.
References
-
- Winch P. Social and cultural responses to emerging vector-borne diseases. J Vector Ecol. 1998;23:47–53. - PubMed
-
- Franz AW, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. - DOI - PMC - PubMed
-
- Alphey L, Beard CB, Billingsley P, Coetzee M, Crisanti A, Curtis C, Eggleston P, Godfray C, Hemingway J, Jacobs-Lorena M, James AA, Kafatos FC, Mukwaya LG, Paton M, Powell JR, Schneider W, Scott TW, Sina B, Sinden R, Sinkins S, Spielman A, Toure Y, Collins FH. Malaria control with genetically manipulated insect vectors. Science. 2002;298:119–121. doi: 10.1126/science.1078278. - DOI - PubMed
-
- O'Brochta DA, Sethuraman N, Wilson R, Hice RH, Pinkerton AC, Levesque CS, Bideshi DK, Jasinskiene N, Coates CJ, James AA, Lehane MJ, Atkinson PW. Gene vector and transposable element behavior in mosquitoes. The Journal of experimental biology. 2003;206:3823–3834. doi: 10.1242/jeb.00638. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

