Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex
- PMID: 18088424
- PMCID: PMC2245944
- DOI: 10.1186/1471-2164-8-464
Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex
Abstract
Background: Sexual reproduction is a core biological function that is conserved throughout eukaryotic evolution, yet breeding systems are extremely variable. Genome-wide comparative studies can be effectively used to identify genes and regulatory patterns that are constrained to preserve core functions from those that may help to account for the diversity of animal reproductive strategies. We use a custom microarray to investigate gene expression in males and two reproductive stages of females in the crustacean Daphnia pulex. Most Daphnia species reproduce by cyclical parthenogenesis, alternating between sexual and clonal reproduction. Both sex determination and the switch in their mode of reproduction is environmentally induced, making Daphnia an interesting comparative system for the study of sex-biased and reproductive genes.
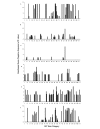
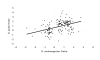
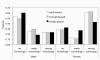
Results: Patterns of gene expression in females and males reveal that 50% of assayed transcripts show some degree of sex-bias. Female-biased transcription is enriched for translation, metabolic and regulatory genes associated with development. Male-biased expression is enriched for cuticle and protease function. Comparison with well studied arthropods such as Drosophila melanogaster and Anopheles gambiae suggests that female-biased patterns tend to be conserved, whereas male-biased genes are evolving faster in D. pulex. These findings are based on the proportion of female-biased, male-biased, and unbiased genes that share sequence similarity with proteins in other animal genomes.
Conclusion: Some transcriptional differences between males and females appear to be conserved across Arthropoda, including the rapid evolution of male-biased genes which is observed in insects and now in a crustacean. Yet, novel patterns of male-biased gene expression are also uncovered. This study is an important first step towards a detailed understanding of the genetic basis and evolution of parthenogenesis, environmental sex determination, and adaptation to aquatic environments.
Figures
Similar articles
-
De Novo Transcriptome Assembly and Sex-Biased Gene Expression in the Cyclical Parthenogenetic Daphnia galeata.Genome Biol Evol. 2016 Oct 23;8(10):3120-3139. doi: 10.1093/gbe/evw221. Genome Biol Evol. 2016. PMID: 27604882 Free PMC article.
-
Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution.BMC Evol Biol. 2009 Apr 21;9:78. doi: 10.1186/1471-2148-9-78. BMC Evol Biol. 2009. PMID: 19383157 Free PMC article.
-
Identification of General Patterns of Sex-Biased Expression in Daphnia, a Genus with Environmental Sex Determination.G3 (Bethesda). 2018 May 4;8(5):1523-1533. doi: 10.1534/g3.118.200174. G3 (Bethesda). 2018. PMID: 29535148 Free PMC article.
-
Genomics of environmentally induced phenotypes in 2 extremely plastic arthropods.J Hered. 2011 Sep-Oct;102(5):512-25. doi: 10.1093/jhered/esr020. Epub 2011 Apr 27. J Hered. 2011. PMID: 21525179 Free PMC article. Review.
-
Environmentally Dependent Alteration of Reproductive Strategies and Juvenile Hormone Signaling in Daphnia (Crustacea: Cladocera).Zoolog Sci. 2025 Feb;42(1). doi: 10.2108/zs240054. Zoolog Sci. 2025. PMID: 39932751 Review.
Cited by
-
Genotypic similarities among the parthenogenetic Darevskia rock lizards with different hybrid origins.BMC Evol Biol. 2020 Sep 16;20(1):122. doi: 10.1186/s12862-020-01690-9. BMC Evol Biol. 2020. PMID: 32938384 Free PMC article.
-
Sexual Transcription Differences in Brachymeria lasus (Hymenoptera: Chalcididae), a Pupal Parasitoid Species of Lymantria dispar (Lepidoptera: Lymantriidae).Front Genet. 2019 Mar 5;10:172. doi: 10.3389/fgene.2019.00172. eCollection 2019. Front Genet. 2019. PMID: 30891067 Free PMC article.
-
CRISPR/Cas-mediated knock-in via non-homologous end-joining in the crustacean Daphnia magna.PLoS One. 2017 Oct 18;12(10):e0186112. doi: 10.1371/journal.pone.0186112. eCollection 2017. PLoS One. 2017. PMID: 29045453 Free PMC article.
-
Comparative Analyses of Reproductive Caste Types Reveal Vitellogenin Genes Involved in Queen Fertility in Solenopsis invicta.Int J Mol Sci. 2023 Dec 5;24(24):17130. doi: 10.3390/ijms242417130. Int J Mol Sci. 2023. PMID: 38138959 Free PMC article.
-
De Novo Transcriptome Assembly and Sex-Biased Gene Expression in the Cyclical Parthenogenetic Daphnia galeata.Genome Biol Evol. 2016 Oct 23;8(10):3120-3139. doi: 10.1093/gbe/evw221. Genome Biol Evol. 2016. PMID: 27604882 Free PMC article.
References
-
- Li Y, Chia J, Bartfai R, Christoffels A, Yue G, Ding K, Ho M, Hill J, Stupka E, Orban L. Comparative analysis of the testis and ovary transcriptomes in zebrafish by combining experimental and computational tools. Comparative and Functional Genomics. 2004;5:403–418. doi: 10.1002/cfg.418. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

