Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs
- PMID: 18089574
- PMCID: PMC2958778
- DOI: 10.1074/jbc.M708734200
Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs
Abstract
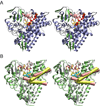
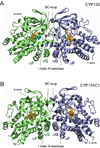
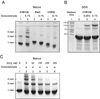
CYP130 is one of the 20 Mycobacterium tuberculosis cytochrome P450 enzymes, only two of which, CYP51 and CYP121, have so far been studied as individually expressed proteins. Here we characterize a third heterologously expressed M. tuberculosis cytochrome P450, CYP130, by UV-visible spectroscopy, isothermal titration calorimetry, and x-ray crystallography, including determination of the crystal structures of ligand-free and econazole-bound CYP130 at a resolution of 1.46 and 3.0A(,) respectively. Ligand-free CYP130 crystallizes in an "open" conformation as a monomer, whereas the econazole-bound form crystallizes in a "closed" conformation as a dimer. Conformational changes enabling the "open-closed" transition involve repositioning of the BC-loop and the F and G helices that envelop the inhibitor in the binding site and reshape the protein surface. Crystal structure analysis shows that the portion of the BC-loop relocates as much as 18A between the open and closed conformations. Binding of econazole to CYP130 involves a conformational change and is mediated by both a set of hydrophobic interactions with amino acid residues in the active site and coordination of the heme iron. CYP130 also binds miconazole with virtually the same binding affinity as econazole and clotrimazole and ketoconazole with somewhat lower affinities, which makes it a plausible target for this class of therapeutic drugs. Overall, binding of the azole inhibitors is a sequential two-step, entropy-driven endothermic process. Binding of econazole and clotrimazole exhibits positive cooperativity that may reflect a propensity of CYP130 to associate into a dimeric structure.
Figures


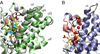

Similar articles
-
An enlarged, adaptable active site in CYP164 family P450 enzymes, the sole P450 in Mycobacterium leprae.Antimicrob Agents Chemother. 2012 Jan;56(1):391-402. doi: 10.1128/AAC.05227-11. Epub 2011 Oct 28. Antimicrob Agents Chemother. 2012. PMID: 22037849 Free PMC article.
-
Structural basis of human CYP51 inhibition by antifungal azoles.J Mol Biol. 2010 Apr 9;397(4):1067-78. doi: 10.1016/j.jmb.2010.01.075. Epub 2010 Feb 10. J Mol Biol. 2010. PMID: 20149798
-
Expression and characterization of Mycobacterium tuberculosis CYP144: common themes and lessons learned in the M. tuberculosis P450 enzyme family.Biochim Biophys Acta. 2011 Jan;1814(1):76-87. doi: 10.1016/j.bbapap.2010.05.015. Epub 2010 Jun 8. Biochim Biophys Acta. 2011. PMID: 20621636
-
CYP121, CYP51 and associated redox systems in Mycobacterium tuberculosis: towards deconvoluting enzymology of P450 systems in a human pathogen.Biochem Soc Trans. 2006 Dec;34(Pt 6):1178-82. doi: 10.1042/BST0341178. Biochem Soc Trans. 2006. PMID: 17073780 Review.
-
Potential drug targets in the Mycobacterium tuberculosis cytochrome P450 system.J Inorg Biochem. 2018 Mar;180:235-245. doi: 10.1016/j.jinorgbio.2018.01.010. Epub 2018 Jan 12. J Inorg Biochem. 2018. PMID: 29352597 Free PMC article. Review.
Cited by
-
Anesthetic drug development: Novel drugs and new approaches.Surg Neurol Int. 2013 Mar 19;4(Suppl 1):S2-S10. doi: 10.4103/2152-7806.109179. Print 2013. Surg Neurol Int. 2013. PMID: 23653886 Free PMC article.
-
The Binary Mixtures of Lambda-Cyhalothrin, Chlorfenapyr, and Abamectin, against the House Fly Larvae, Musca domestica L.Molecules. 2022 May 12;27(10):3084. doi: 10.3390/molecules27103084. Molecules. 2022. PMID: 35630573 Free PMC article.
-
Identification, characterization, and azole-binding properties of Mycobacterium smegmatis CYP164A2, a homolog of ML2088, the sole cytochrome P450 gene of Mycobacterium leprae.Antimicrob Agents Chemother. 2009 Mar;53(3):1157-64. doi: 10.1128/AAC.01237-08. Epub 2008 Dec 15. Antimicrob Agents Chemother. 2009. PMID: 19075057 Free PMC article.
-
Deletion of the Mycobacterium tuberculosis cyp138 gene leads to changes in membrane-related lipid composition and antibiotic susceptibility.Front Microbiol. 2024 Mar 25;15:1301204. doi: 10.3389/fmicb.2024.1301204. eCollection 2024. Front Microbiol. 2024. PMID: 38591032 Free PMC article.
-
Interactions of cytochrome P450s with their ligands.Arch Biochem Biophys. 2011 Mar 1;507(1):56-65. doi: 10.1016/j.abb.2010.10.006. Epub 2010 Oct 19. Arch Biochem Biophys. 2011. PMID: 20939998 Free PMC article. Review.
References
-
- Zhang Y. Annu. Rev. Pharmacol. Toxicol. 2005;45:529–564. - PubMed
-
- Ahmad Z, Sharma S, Khuller GK. FEMS Microbiology Letters. 2005;251:19–22. - PubMed
-
- Ahmad Z, Sharma S, Khuller GK, Singh P, Faujdar J, Katoch VM. Int. J. Antimicrob. Agents. 2006;28:543–544. - PubMed
-
- Ahmad Z, Sharma S, Khuller GK. FEMS Microbiology Letters. 2006;261:181–186. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

