A map of human cancer signaling
- PMID: 18091723
- PMCID: PMC2174632
- DOI: 10.1038/msb4100200
A map of human cancer signaling
Abstract
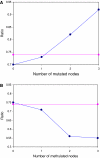
We conducted a comprehensive analysis of a manually curated human signaling network containing 1634 nodes and 5089 signaling regulatory relations by integrating cancer-associated genetically and epigenetically altered genes. We find that cancer mutated genes are enriched in positive signaling regulatory loops, whereas the cancer-associated methylated genes are enriched in negative signaling regulatory loops. We further characterized an overall picture of the cancer-signaling architectural and functional organization. From the network, we extracted an oncogene-signaling map, which contains 326 nodes, 892 links and the interconnections of mutated and methylated genes. The map can be decomposed into 12 topological regions or oncogene-signaling blocks, including a few 'oncogene-signaling-dependent blocks' in which frequently used oncogene-signaling events are enriched. One such block, in which the genes are highly mutated and methylated, appears in most tumors and thus plays a central role in cancer signaling. Functional collaborations between two oncogene-signaling-dependent blocks occur in most tumors, although breast and lung tumors exhibit more complex collaborative patterns between multiple blocks than other cancer types. Benchmarking two data sets derived from systematic screening of mutations in tumors further reinforced our findings that, although the mutations are tremendously diverse and complex at the gene level, clear patterns of oncogene-signaling collaborations emerge recurrently at the network level. Finally, the mutated genes in the network could be used to discover novel cancer-associated genes and biomarkers.
Figures


References
-
- Awan A, Bari H, Yan F, Mokin S, Yang S, Chowdhury Q, Yu Z, Purisima EO, Wang E (2007) Regulatory network motifs and hotspots of cancer genes in a mammalian cellular signaling network. IET Syst Biol 1: 292–297 - PubMed
-
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004) Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 - PubMed
-
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, van de RM, Brown PO, van d V (2005) Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci USA 102: 3738–3743 - PMC - PubMed
-
- Chanock SJ, Thomas G (2007) The devil is in the DNA. Nat Genet 39: 283–284 - PubMed
-
- Chng WJ (2007) Limits to the human cancer genome project? Science 315: 762–765 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

