Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability
- PMID: 18093314
- PMCID: PMC2225408
- DOI: 10.1186/1471-2091-8-27
Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability
Abstract
Background: The COP9 signalosome (CSN) is a conserved protein complex in eukaryotic cells consisting of eight subunits (CSN1 to CSN8). Recent data demonstrate that the CSN is a regulator of the ubiquitin (Ub) proteasome system (UPS). It controls substrate ubiquitination by cullin-RING Ub ligases (CRLs), a process that determines substrate specificity of the UPS. The intrinsic deneddylating activity localized to CSN5 as well as the associated kinases and deubiquitinating activity are involved in the regulatory function of CSN. The exact mechanisms are unclear. In this study we knocked down CSN1 (siCSN1), CSN3 (siCSN3) and CSN5 (siCSN5) by specific siRNA oligos permanently expressed in HeLa cells. The analysis and comparison of siRNA cells revealed differential impact of individual subunits on CSN structure and function.
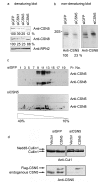
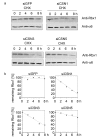
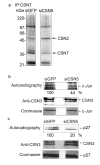
Results: Permanent knockdowns of CSN1 and CSN3 led to a reduction of the subunits to approximately 40%, which is accompanied by a proportional decrease of the CSN holocomplex. In contrast, downregulation of CSN5 in HeLa cells reduced the CSN5 protein below 20% without significant effects on the remaining complex. The CRL component Rbx1 was characterized by accelerated proteolysis in siCSN1 and siCSN3 and also in siCSN5 cells, however, with lesser extent. Immunoprecipitated CSN complex from siCSN5 cells was less effective in phosphorylating c-Jun and p27. Accelerated degradation of c-Jun in siCSN5 cells was rescued by overexpression of CSN5 as well as of the deneddylation mutant CSN5D151N. Overexpression of CSN5 cannot rescue c-Jun destabilization in siCSN1.
Conclusion: There exists a coordinated downregulation of CSN subunits in the CSN1 and CSN3 knockdowns. The underlying regulatory mechanisms are obscure. CSN5 seems to possess a specific status in HeLa cells. Its reduction is not connected with coordinated downregulation of other subunits. CSN knockdowns confirm that the stabilization of the CRL component Rbx1 is a major CSN function. In addition, downregulation of CSN subunits influences the stability of important cellular regulators such as c-Jun and p27.
Figures


References
-
- Bech-Otschir D, Kapelari B, Dubiel W. The COP9 Signalosome: Its Possible Role in the Ubiquitin System. In: Mayer R, Ciechanover A, Rechsteiner M, editor. Protein Degradation. 1: Ubiquitin and the Chemistry of Life. Weinheim , WILEY-VCH Verlag GmbH & Co. KGaA; 2005. pp. 348–369.
-
- Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–286. - PubMed
-
- Deng XW, Dubiel W, Wei N, Hofmann K, Mundt K, Colicelli J, Kato J, Naumann M, Segal D, Seeger M, Carr A, Glickman M, Chamovitz DA. Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet. 2000;16:202–203. - PubMed
-
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

