Checkpoint independence of most DNA replication origins in fission yeast
- PMID: 18093330
- PMCID: PMC2235891
- DOI: 10.1186/1471-2199-8-112
Checkpoint independence of most DNA replication origins in fission yeast
Abstract
Background: In budding yeast, the replication checkpoint slows progress through S phase by inhibiting replication origin firing. In mammals, the replication checkpoint inhibits both origin firing and replication fork movement. To find out which strategy is employed in the fission yeast, Schizosaccharomyces pombe, we used microarrays to investigate the use of origins by wild-type and checkpoint-mutant strains in the presence of hydroxyurea (HU), which limits the pool of deoxyribonucleoside triphosphates (dNTPs) and activates the replication checkpoint. The checkpoint-mutant cells carried deletions either of rad3 (which encodes the fission yeast homologue of ATR) or cds1 (which encodes the fission yeast homologue of Chk2).
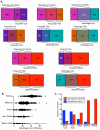
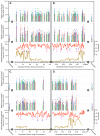
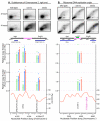
Results: Our microarray results proved to be largely consistent with those independently obtained and recently published by three other laboratories. However, we were able to reconcile differences between the previous studies regarding the extent to which fission yeast replication origins are affected by the replication checkpoint. We found (consistent with the three previous studies after appropriate interpretation) that, in surprising contrast to budding yeast, most fission yeast origins, including both early- and late-firing origins, are not significantly affected by checkpoint mutations during replication in the presence of HU. A few origins (approximately 3%) behaved like those in budding yeast: they replicated earlier in the checkpoint mutants than in wild type. These were located primarily in the heterochromatic subtelomeric regions of chromosomes 1 and 2. Indeed, the subtelomeric regions defined by the strongest checkpoint restraint correspond precisely to previously mapped subtelomeric heterochromatin. This observation implies that subtelomeric heterochromatin in fission yeast differs from heterochromatin at centromeres, in the mating type region, and in ribosomal DNA, since these regions replicated at least as efficiently in wild-type cells as in checkpoint-mutant cells.
Conclusion: The fact that approximately 97% of fission yeast replication origins - both early and late - are not significantly affected by replication checkpoint mutations in HU-treated cells suggests that (i) most late-firing origins are restrained from firing in HU-treated cells by at least one checkpoint-independent mechanism, and (ii) checkpoint-dependent slowing of S phase in fission yeast when DNA is damaged may be accomplished primarily by the slowing of replication forks.
Figures






Similar articles
-
Checkpoint effects and telomere amplification during DNA re-replication in fission yeast.BMC Mol Biol. 2007 Dec 21;8:119. doi: 10.1186/1471-2199-8-119. BMC Mol Biol. 2007. PMID: 18154680 Free PMC article.
-
Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast.EMBO J. 2007 Mar 7;26(5):1327-39. doi: 10.1038/sj.emboj.7601585. Epub 2007 Feb 15. EMBO J. 2007. PMID: 17304213 Free PMC article.
-
Regulation of DNA replication machinery by Mrc1 in fission yeast.Genetics. 2006 Sep;174(1):155-65. doi: 10.1534/genetics.106.060053. Epub 2006 Jul 18. Genetics. 2006. PMID: 16849602 Free PMC article.
-
Checkpoint functions of RecQ helicases at perturbed DNA replication fork.Curr Genet. 2021 Jun;67(3):369-382. doi: 10.1007/s00294-020-01147-y. Epub 2021 Jan 11. Curr Genet. 2021. PMID: 33427950 Review.
-
Controlling DNA replication origins in response to DNA damage - inhibit globally, activate locally.J Cell Sci. 2013 Mar 15;126(Pt 6):1297-306. doi: 10.1242/jcs.096701. J Cell Sci. 2013. PMID: 23645160 Review.
Cited by
-
Genome-wide identification and characterization of replication origins by deep sequencing.Genome Biol. 2012 Apr 24;13(4):R27. doi: 10.1186/gb-2012-13-4-r27. Genome Biol. 2012. PMID: 22531001 Free PMC article.
-
ATR: an essential regulator of genome integrity.Nat Rev Mol Cell Biol. 2008 Aug;9(8):616-27. doi: 10.1038/nrm2450. Epub 2008 Jul 2. Nat Rev Mol Cell Biol. 2008. PMID: 18594563 Free PMC article. Review.
-
Genome-wide estimation of firing efficiencies of origins of DNA replication from time-course copy number variation data.BMC Bioinformatics. 2010 May 13;11:247. doi: 10.1186/1471-2105-11-247. BMC Bioinformatics. 2010. PMID: 20462459 Free PMC article.
-
Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres.EMBO J. 2009 Apr 8;28(7):810-20. doi: 10.1038/emboj.2009.31. Epub 2009 Feb 12. EMBO J. 2009. PMID: 19214192 Free PMC article.
-
Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels.J Cell Biol. 2010 Jan 25;188(2):209-21. doi: 10.1083/jcb.200911037. Epub 2010 Jan 18. J Cell Biol. 2010. PMID: 20083602 Free PMC article.
References
-
- Lee JK, Moon KY, Jiang Y, Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc Natl Acad Sci USA. 2001;98:13589–13594. doi: 10.1073/pnas.251530398. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

