Continuity conditions and torsion angles from ssNMR orientational restraints
- PMID: 18093855
- PMCID: PMC2435099
- DOI: 10.1016/j.jmr.2007.11.018
Continuity conditions and torsion angles from ssNMR orientational restraints
Abstract
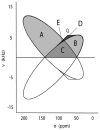
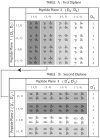
The backbone torsion angle pair (varphi,psi) at each amino acid of a polypeptide is a descriptor of its conformation. One can use chemical shift and dipolar coupling data from solid-state NMR PISEMA experiments to directly calculate the torsion angles for the membrane-spanning portion of a protein. However, degeneracies inherent in the data give rise to multiple potential torsion angles between two adjacent peptide planes (a diplane). The molecular backbone structure can be determined by gluing together the consecutive diplanes, as in the PIPATH algorithm [T. Asbury, J.R. Quine, S. Achuthan, J. Hu, M.S. Chapman, T.A. Cross, R. Bertram, PIPATH: an optimized alogrithm for generating alpha-helical structures from PISEMA data, J. Magn. Reson. 183 (2006) 87-95.]. The multiplicities in torsion angles translate to multiplicities in diplane orientations. In this paper, we show that adjacent diplanes can be glued together to form a permissible structure only if they satisfy continuity conditions, described quantitatively here. These restrict the number of potential torsion angle pairs. We rewrite the torsion angle formulas from [J.R. Quine, M.T. Brenneman, T.A. Cross, Protein structural analysis from solid-state NMR-drived orientational constraints, Biophys. J. 72 (1997) 2342-2348.] so that they automatically satisfy the continuity conditions. The reformulated torsion angle formulas have been applied recently in the PIPATH algorithm [T. Asbury, J.R. Quine, S. Achuthan, J. Hu, M.S. Chapman, T.A. Cross, R. Bertram, PIPATH: an optimized alogrithm for generating alpha-helical structures from PISEMA data, J. Magn. Reson. 183 (2006) 87-95.] and will be helpful in other applications in which diplane gluing is used to construct a protein backbone model.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

