Combinatorics of feedback in cellular uptake and metabolism of small molecules
- PMID: 18093927
- PMCID: PMC2409224
- DOI: 10.1073/pnas.0706231105
Combinatorics of feedback in cellular uptake and metabolism of small molecules
Abstract
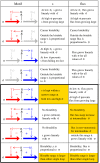
We analyze the connection between structure and function for regulatory motifs associated with cellular uptake and usage of small molecules. Based on the boolean logic of the feedback we suggest four classes: the socialist, consumer, fashion, and collector motifs. We find that the socialist motif is good for homeostasis of a useful but potentially poisonous molecule, whereas the consumer motif is optimal for nutrition molecules. Accordingly, examples of these motifs are found in, respectively, the iron homeostasis system in various organisms and in the uptake of sugar molecules in bacteria. The remaining two motifs have no obvious analogs in small molecule regulation, but we illustrate their behavior using analogies to fashion and obesity. These extreme motifs could inspire construction of synthetic systems that exhibit bistable, history-dependent states, and homeostasis of flux (rather than concentration).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Dynamics of uptake and metabolism of small molecules in cellular response systems.PLoS One. 2009;4(3):e4923. doi: 10.1371/journal.pone.0004923. Epub 2009 Mar 17. PLoS One. 2009. PMID: 19290058 Free PMC article.
-
Damped oscillations in the adaptive response of the iron homeostasis network of E. coli.Mol Microbiol. 2010 Apr;76(2):428-36. doi: 10.1111/j.1365-2958.2010.07111.x. Epub 2010 Mar 16. Mol Microbiol. 2010. PMID: 20345668
-
Environmental selection of the feed-forward loop circuit in gene-regulation networks.Phys Biol. 2005 Jun;2(2):81-8. doi: 10.1088/1478-3975/2/2/001. Phys Biol. 2005. PMID: 16204860
-
Simplified models of biological networks.Annu Rev Biophys. 2010;39:43-59. doi: 10.1146/annurev.biophys.093008.131241. Annu Rev Biophys. 2010. PMID: 20192769 Review.
-
Iron transport in Escherichia coli: all has not been said and done.Biometals. 2006 Apr;19(2):159-72. doi: 10.1007/s10534-005-4341-2. Biometals. 2006. PMID: 16718601 Review.
Cited by
-
Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli.Nat Commun. 2014 Sep 15;5:4910. doi: 10.1038/ncomms5910. Nat Commun. 2014. PMID: 25222563 Free PMC article.
-
Dynamics of uptake and metabolism of small molecules in cellular response systems.PLoS One. 2009;4(3):e4923. doi: 10.1371/journal.pone.0004923. Epub 2009 Mar 17. PLoS One. 2009. PMID: 19290058 Free PMC article.
-
Deciphering the transcriptional regulatory logic of amino acid metabolism.Nat Chem Biol. 2011 Nov 13;8(1):65-71. doi: 10.1038/nchembio.710. Nat Chem Biol. 2011. PMID: 22082910 Free PMC article.
-
Relation of intracellular signal levels and promoter activities in the gal regulon of Escherichia coli.J Mol Biol. 2009 Aug 28;391(4):671-8. doi: 10.1016/j.jmb.2009.06.043. Epub 2009 Jun 23. J Mol Biol. 2009. PMID: 19559028 Free PMC article.
-
Growth landscape formed by perception and import of glucose in yeast.Nature. 2009 Dec 17;462(7275):875-9. doi: 10.1038/nature08653. Nature. 2009. PMID: 20016593 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

