Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends
- PMID: 18093953
- PMCID: PMC2409239
- DOI: 10.1073/pnas.0708541104
Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends
Abstract
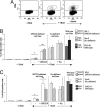
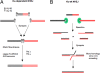
XRCC4-null mice have a more severe phenotype than KU80-null mice. Here, we address whether this difference in phenotype is connected to nonhomologous end-joining (NHEJ). We used intrachromosomal substrates to monitor NHEJ of two distal double-strand breaks (DSBs) targeted by I-SceI, in living cells. In xrcc4-defective XR-1 cells, a residual but significant end-joining process exists, which primarily uses microhomologies distal from the DSB. However, NHEJ efficiency was strongly reduced in xrcc4-defective XR-1 cells versus complemented cells, contrasting with KU-deficient xrs6 cells, which showed levels of end-joining similar to those of complemented cells. Nevertheless, sequence analysis of the repair junctions indicated that the accuracy of end-joining was strongly affected in both xrcc4-deficient and KU-deficient cells. More specifically, these data showed that the KU80/XRCC4 pathway is conservative and not intrinsically error-prone but can accommodate non-fully complementary ends at the cost of limited mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Alternative endings.Proc Natl Acad Sci U S A. 2008 Jan 15;105(2):405-6. doi: 10.1073/pnas.0711334105. Epub 2008 Jan 7. Proc Natl Acad Sci U S A. 2008. PMID: 18180452 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

