DYC-1, a protein functionally linked to dystrophin in Caenorhabditis elegans is associated with the dense body, where it interacts with the muscle LIM domain protein ZYX-1
- PMID: 18094057
- PMCID: PMC2262962
- DOI: 10.1091/mbc.e07-05-0497
DYC-1, a protein functionally linked to dystrophin in Caenorhabditis elegans is associated with the dense body, where it interacts with the muscle LIM domain protein ZYX-1
Abstract
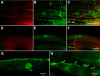
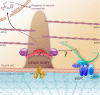
In Caenorhabditis elegans, mutations of the dystrophin homologue, dys-1, produce a peculiar behavioral phenotype (hyperactivity and a tendency to hypercontract). In a sensitized genetic background, dys-1 mutations also lead to muscle necrosis. The dyc-1 gene was previously identified in a genetic screen because its mutation leads to the same phenotype as dys-1, suggesting that the two genes are functionally linked. Here, we report the detailed characterization of the dyc-1 gene. dyc-1 encodes two isoforms, which are expressed in neurons and muscles. Isoform-specific RNAi experiments show that the absence of the muscle isoform, and not that of the neuronal isoform, is responsible for the dyc-1 mutant phenotype. In the sarcomere, the DYC-1 protein is localized at the edges of the dense body, the nematode muscle adhesion structure where actin filaments are anchored and linked to the sarcolemma. In yeast two-hybrid assays, DYC-1 interacts with ZYX-1, the homologue of the vertebrate focal adhesion LIM domain protein zyxin. ZYX-1 localizes at dense bodies and M-lines as well as in the nucleus of C. elegans striated muscles. The DYC-1 protein possesses a highly conserved 19 amino acid sequence, which is involved in the interaction with ZYX-1 and which is sufficient for addressing DYC-1 to the dense body. Altogether our findings indicate that DYC-1 may be involved in dense body function and stability. This, taken together with the functional link between the C. elegans DYC-1 and DYS-1 proteins, furthermore suggests a requirement of dystrophin function at this structure. As the dense body shares functional similarity with both the vertebrate Z-disk and the costamere, we therefore postulate that disruption of muscle cell adhesion structures might be the primary event of muscle degeneration occurring in the absence of dystrophin, in C. elegans as well as vertebrates.
Figures







References
-
- Ahn A. H., Kunkel L. M. The structural and functional diversity of dystrophin. Nat. Genet. 1993;3:283–291. - PubMed
-
- Bessou C., Giugia J. B., Franks C. J., Holden-Dye L., Segalat L. Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission. Neurogenetics. 1998;2:61–72. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

