Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site
- PMID: 18094123
- PMCID: PMC2212249
- DOI: 10.1261/rna.696508
Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site
Abstract
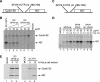
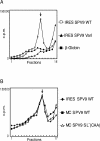
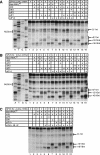
The Simian picornavirus type 9 (SPV9) 5'-untranslated region (5' UTR) has been predicted to contain an internal ribosomal entry site (IRES) with structural elements that resemble domains of hepacivirus/pestivirus (HP) IRESs. In vitro reconstitution of initiation confirmed that this 5' UTR contains an IRES and revealed that it has both functional similarities and differences compared to HP IRESs. Like HP IRESs, the SPV9 IRES bound directly to 40S subunits and eukaryotic initiation factor (eIF) 3, depended on the conserved domain IIId for ribosomal binding and consequently for function, and additionally required eIF2/initiator tRNA to yield 48S complexes that formed elongation-competent 80S ribosomes in the presence of eIF5, eIF5B, and 60S subunits. Toeprinting analysis revealed that eIF1A stabilized 48S complexes, whereas eIF1 induced conformational changes in the 40S subunit, likely corresponding to partial opening of the entry latch of the mRNA-binding channel, that were exacerbated by eIF3 and suppressed by eIF1A. The SPV9 IRES differed from HP IRESs in that its function was enhanced by eIF4A/eIF4F when the IRES was adjacent to the wild-type coding sequence, but was less affected by these factors or by a dominant negative eIF4A mutant when potentially less structured coding sequences were present. Exceptionally, this IRES promoted binding of initiator tRNA to the initiation codon in the P site of 40S subunits independently of eIF2. Although these 40S/IRES/tRNA complexes could not form active 80S ribosomes, this constitutes a second difference between the SPV9 and HP IRESs. eIF1 destabilized the eIF2-independent ribosomal binding of initiator tRNA.
Figures






References
-
- Algire, M.A., Maag, D., Lorsch, J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. - PubMed
-
- Alkalaeva, E.Z., Pisarev, A.V., Frolova, L.Y., Kisselev, L.L., Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. - PubMed
-
- Battiste, J.L., Pestova, T.V., Hellen, C.U.T., Wagner, G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. - PubMed
-
- Bordeleau, M.E., Mori, A., Oberer, M., Lindqvist, L., Chard, L.S., Higa, T., Belsham, G.J., Wagner, G., Tanaka, J., Pelletier, J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. - PubMed
-
- Carter, A.P., Clemons W.M., Jr, Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., Ramakrishnan, V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
