Polymerization properties of the Thermotoga maritima actin MreB: roles of temperature, nucleotides, and ions
- PMID: 18095710
- PMCID: PMC2562234
- DOI: 10.1021/bi701538e
Polymerization properties of the Thermotoga maritima actin MreB: roles of temperature, nucleotides, and ions
Abstract
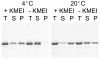
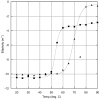
MreB is a bacterial orthologue of actin that affects cell shape, polarity, and chromosome segregation. Although a significant body of work has explored its cellular functions, we know very little about the biochemical behavior of MreB. We have cloned, overexpressed in Escherichia coli, and purified untagged MreB1 from Thermotoga maritima. We have characterized the conditions that regulate its monomer-to-polymer assembly reaction, the critical concentrations of that reaction, the manner in which MreB uses nucleotides, its stability, and the structure of the assembled polymer. MreB requires a bound purine nucleotide for polymerization and rapidly hydrolyzes it following assembly. MreB assembly contains two distinct components, one that does not require divalent cations and one that does, which may comprise the nucleation and elongation phases of assembly, respectively. MreB assembly is strongly favored by increasing temperature or protein concentration but inhibited differentially by high concentrations of monovalent salts. The polymerization rate increases and the bulk critical concentration decreases with increasing temperature, but in contrast to previous reports, MreB is capable of polymerizing across a broad range of temperatures. MreB polymers are shorter and stiffer and scatter more light than eukaryotic actin filaments. Due to rapid ATP hydrolysis and phosphate release, we suggest that most assembled MreB in cells is in the ADP-bound state. Because of only moderate differences between the ATP and ADP critical concentrations, treadmilling may occur, but we do not predict dynamic instability in cells. Because of the relatively low cellular concentration of MreB and the observed structural properties of the polymer, a single MreB assembly may exist in cells.
Figures














References
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular Biology of the Cell - Second Edition. Garland; New York: 1989.
-
- Fitch W. Distinguishing homologous from analogous proteins. Systematic Zoology. 1970;19:99–113. - PubMed
-
- Koonin EV. Orthologs, Paralogs, and Evolutionary Genomics. Annual Review of Genetics. 2005;39:309–338. - PubMed
-
- Jones LJF, Carballido-Lopez R, Errington J. Control of Cell Shape in Bacteria: Helical, Actin-like Filaments in Bacillus subtilis. Cell. 2001;104:913–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

