Fibroblast activation protein peptide substrates identified from human collagen I derived gelatin cleavage sites
- PMID: 18095711
- PMCID: PMC4696028
- DOI: 10.1021/bi701921b
Fibroblast activation protein peptide substrates identified from human collagen I derived gelatin cleavage sites
Abstract
A highly consistent trait of tumor stromal fibroblasts is the induction of the membrane-bound serine protease fibroblast activation protein-alpha (FAP), which is overexpressed on the surface of reactive stromal fibroblasts present within the stroma of the majority of human epithelial tumors. In contrast, FAP is not expressed by tumor epithelial cells or by fibroblasts or other cell types in normal tissues. The proteolytic activity of FAP, therefore, represents a potential pan-tumor target that can be exploited for the release of potent cytotoxins from inactive prodrugs consisting of an FAP peptide substrate coupled to a cytotoxin. To identify FAP peptide substrates, we used liquid chromatography tandem mass spectroscopy based sequencing to generate a complete map of the FAP cleavage sites within human collagen I derived gelatin. Positional analysis of the frequency of each amino acid at each position within the cleavage sites revealed FAP consensus sequences PPGP and (D/E)-(R/K)-G-(E/D)-(T/S)-G-P. These studies further demonstrated that ranking cleavage sites based on the magnitude of the LC/MS/MS extracted ion current predicted FAP substrates that were cleaved with highest efficiency. Fluorescence-quenched peptides were synthesized on the basis of the cleavage sites with the highest ion current rankings, and kinetic parameters for FAP hydrolysis were determined. The substrate DRGETGP, which corresponded to the consensus sequence, had the lowest Km of 21 microM. Overall the Km values were relatively similar for both high and low ranked substrates, whereas the kcat values differed by up to 100-fold. On the basis of these results, the FAP consensus sequences are currently being evaluated as FAP-selective peptide carriers for incorporation into FAP-activated prodrugs.
Figures

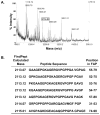

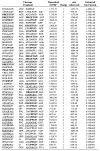

References
-
- Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. - PubMed
-
- Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64:327–336. - PubMed
-
- Basset P, Bellocq JP, Wolf C, Stoll I, Hutin P, Limacher JM, Podhajcer OL, Chenard MP, Rio MC, Chambon P. A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature. 1990;348:699–704. - PubMed
-
- Brown LF, Guidi AJ, Schnitt SJ, Van De Water L, Iruela-Arispe ML, Yeo TK, Tognazzi K, Dvorak HF. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin Cancer Res. 1999;5:1041–1056. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

