CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction
- PMID: 18096039
- PMCID: PMC2246271
- DOI: 10.1186/gb-2007-8-12-r269
CONTRAST: a discriminative, phylogeny-free approach to multiple informant de novo gene prediction
Abstract
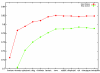
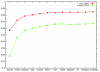
We describe CONTRAST, a gene predictor which directly incorporates information from multiple alignments rather than employing phylogenetic models. This is accomplished through the use of discriminative machine learning techniques, including a novel training algorithm. We use a two-stage approach, in which a set of binary classifiers designed to recognize coding region boundaries is combined with a global model of gene structure. CONTRAST predicts exact coding region structures for 65% more human genes than the previous state-of-the-art method, misses 46% fewer exons and displays comparable gains in specificity.
Figures




References
-
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. - PubMed
-
- Bafna V, Huson DH. The conserved exon method for gene finding. Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology. 2000. pp. 3–12. - PubMed
-
- Korf I, Flicek P, Duan D, Brent MR. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17(Suppl 1):S140–S149. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

