The fungus Ustilago maydis and humans share disease-related proteins that are not found in Saccharomyces cerevisiae
- PMID: 18096044
- PMCID: PMC2262911
- DOI: 10.1186/1471-2164-8-473
The fungus Ustilago maydis and humans share disease-related proteins that are not found in Saccharomyces cerevisiae
Abstract
Background: The corn smut fungus Ustilago maydis is a well-established model system for molecular phytopathology. In addition, it recently became evident that U. maydis and humans share proteins and cellular processes that are not found in the standard fungal model Saccharomyces cerevisiae. This prompted us to do a comparative analysis of the predicted proteome of U. maydis, S. cerevisiae and humans.
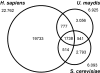
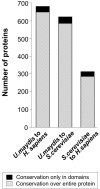
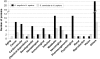
Results: At a cut off at 20% identity over protein length, all three organisms share 1738 proteins, whereas both fungi share only 541 conserved proteins. Despite the evolutionary distance between U. maydis and humans, 777 proteins were shared. When applying a more stringent criterion (> or = 20% identity with a homologue in one organism over at least 50 amino acids and > or = 10% less in the other organism), we found 681 proteins for the comparison of U. maydis and humans, whereas the both fungi share only 622 fungal specific proteins. Finally, we found that S. cerevisiae and humans shared 312 proteins. In the U. maydis to H. sapiens homology set 454 proteins are functionally classified and 42 proteins are related to serious human diseases. However, a large portion of 222 proteins are of unknown function.
Conclusion: The fungus U. maydis has a long history of being a model system for understanding DNA recombination and repair, as well as molecular plant pathology. The identification of functionally un-characterized genes that are conserved in humans and U. maydis opens the door for experimental work, which promises new insight in the cell biology of the mammalian cell.
Figures


References
-
- Evans CS, Hedger JN. Degradation of plant cell wall polymers. In: Gadd GM, editor. Fungi in bioremediation. Cambridge, United kingdom: Cambridge University Press; 2001. pp. 1–26.
-
- Smith SE, Read DJ. Mycorrhizal symbiosis. New York: Academic Press, Inc; 1997.
-
- San-Blas G, Travassos LR, Fries BC, Goldman DL, Casadevall A, Carmona AK, Barros TF, Puccia R, Hostetter MK, Shanks SG, et al. Fungal morphogenesis and virulence. Med Mycol. 2000;38:79–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

