Surface antigens and potential virulence factors from parasites detected by comparative genomics of perfect amino acid repeats
- PMID: 18096064
- PMCID: PMC2254594
- DOI: 10.1186/1477-5956-5-20
Surface antigens and potential virulence factors from parasites detected by comparative genomics of perfect amino acid repeats
Abstract
Background: Many parasitic organisms, eukaryotes as well as bacteria, possess surface antigens with amino acid repeats. Making up the interface between host and pathogen such repetitive proteins may be virulence factors involved in immune evasion or cytoadherence. They find immunological applications in serodiagnostics and vaccine development. Here we use proteins which contain perfect repeats as a basis for comparative genomics between parasitic and free-living organisms.
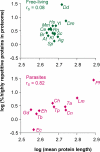
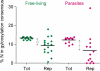
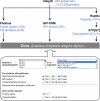
Results: We have developed Reptile http://reptile.unibe.ch, a program for proteome-wide probabilistic description of perfect repeats in proteins. Parasite proteomes exhibited a large variance regarding the proportion of repeat-containing proteins. Interestingly, there was a good correlation between the percentage of highly repetitive proteins and mean protein length in parasite proteomes, but not at all in the proteomes of free-living eukaryotes. Reptile combined with programs for the prediction of transmembrane domains and GPI-anchoring resulted in an effective tool for in silico identification of potential surface antigens and virulence factors from parasites.
Conclusion: Systemic surveys for perfect amino acid repeats allowed basic comparisons between free-living and parasitic organisms that were directly applicable to predict proteins of serological and parasitological importance. An on-line tool is available at http://genomics.unibe.ch/dora.
Figures

Similar articles
-
Repeat-enriched proteins are related to host cell invasion and immune evasion in parasitic protozoa.Mol Biol Evol. 2013 Apr;30(4):951-63. doi: 10.1093/molbev/mst001. Epub 2013 Jan 8. Mol Biol Evol. 2013. PMID: 23303306
-
RepSeq--a database of amino acid repeats present in lower eukaryotic pathogens.BMC Bioinformatics. 2007 Apr 11;8:122. doi: 10.1186/1471-2105-8-122. BMC Bioinformatics. 2007. PMID: 17428323 Free PMC article.
-
COPASAAR--a database for proteomic analysis of single amino acid repeats.BMC Bioinformatics. 2005 Aug 3;6:196. doi: 10.1186/1471-2105-6-196. BMC Bioinformatics. 2005. PMID: 16078990 Free PMC article.
-
Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes.Int J Parasitol. 2008 Jan;38(1):1-31. doi: 10.1016/j.ijpara.2007.07.018. Epub 2007 Sep 15. Int J Parasitol. 2008. PMID: 17949725 Review.
-
Molecular basis for evasion of host immunity and pathogenesis in malaria.Biochim Biophys Acta. 1998 Feb 27;1406(1):10-27. doi: 10.1016/s0925-4439(97)00078-1. Biochim Biophys Acta. 1998. PMID: 9545516 Review.
Cited by
-
Trichomonas vaginalis vast BspA-like gene family: evidence for functional diversity from structural organisation and transcriptomics.BMC Genomics. 2010 Feb 8;11:99. doi: 10.1186/1471-2164-11-99. BMC Genomics. 2010. PMID: 20144183 Free PMC article.
-
Down Modulation of Host Immune Response by Amino Acid Repeats Present in a Trypanosoma cruzi Ribosomal Antigen.Front Microbiol. 2017 Nov 10;8:2188. doi: 10.3389/fmicb.2017.02188. eCollection 2017. Front Microbiol. 2017. PMID: 29176965 Free PMC article.
-
Genome-wide survey and analysis of microsatellites in nematodes, with a focus on the plant-parasitic species Meloidogyne incognita.BMC Genomics. 2010 Oct 25;11:598. doi: 10.1186/1471-2164-11-598. BMC Genomics. 2010. PMID: 20973953 Free PMC article.
-
Reduction and expansion in microsporidian genome evolution: new insights from comparative genomics.Genome Biol Evol. 2013;5(12):2285-303. doi: 10.1093/gbe/evt184. Genome Biol Evol. 2013. PMID: 24259309 Free PMC article.
-
The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution.PLoS Pathog. 2012;8(10):e1002979. doi: 10.1371/journal.ppat.1002979. Epub 2012 Oct 25. PLoS Pathog. 2012. PMID: 23133373 Free PMC article.
References
LinkOut - more resources
Full Text Sources

