Gene prediction: compare and CONTRAST
- PMID: 18096089
- PMCID: PMC2246255
- DOI: 10.1186/gb-2007-8-12-233
Gene prediction: compare and CONTRAST
Abstract
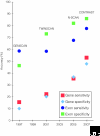
CONTRAST, a new gene-prediction algorithm that uses sophisticated machine-learning techniques, has pushed de novo prediction accuracy to new heights, and has significantly closed the gap between de novo and evidence-based methods for human genome annotation.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

