Activated glucocorticoid receptor interacts with the INHAT component Set/TAF-Ibeta and releases it from a glucocorticoid-responsive gene promoter, relieving repression: implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation
- PMID: 18096310
- PMCID: PMC2350211
- DOI: 10.1016/j.mce.2007.10.014
Activated glucocorticoid receptor interacts with the INHAT component Set/TAF-Ibeta and releases it from a glucocorticoid-responsive gene promoter, relieving repression: implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation
Abstract
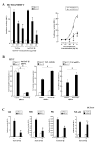
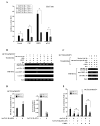
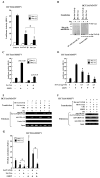
Set/template-activating factor (TAF)-Ibeta, part of the Set-Can oncogene product found in acute undifferentiated leukemia, is a component of the inhibitor of acetyltransferases (INHAT) complex. Set/TAF-Ibeta interacted with the DNA-binding domain of the glucocorticoid receptor (GR) in yeast two-hybrid screening, and repressed GR-induced transcriptional activity of a chromatin-integrated glucocorticoid-responsive and a natural promoter. Set/TAF-Ibeta was co-precipitated with glucocorticoid response elements (GREs) of these promoters in the absence of dexamethasone, while addition of the hormone caused dissociation of Set/TAF-Ibeta from and attraction of the p160-type coactivator GRIP1 to the promoter GREs. Set-Can fusion protein, on the other hand, did not interact with GR, was constitutively co-precipitated with GREs and suppressed GRIP1-induced enhancement of GR transcriptional activity and histone acetylation. Thus, Set/TAF-Ibeta acts as a ligand-activated GR-responsive transcriptional repressor, while Set-Can does not retain physiologic responsiveness to ligand-bound GR, possibly contributing to the poor responsiveness of Set-Can-harboring leukemic cells to glucocorticoids.
Figures



Similar articles
-
Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription.Mol Endocrinol. 2006 Feb;20(2):335-47. doi: 10.1210/me.2005-0248. Epub 2005 Sep 29. Mol Endocrinol. 2006. PMID: 16195249
-
A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression.J Biol Chem. 2004 Jul 16;279(29):30850-5. doi: 10.1074/jbc.M404969200. Epub 2004 May 10. J Biol Chem. 2004. PMID: 15136563
-
Highly acidic C-terminal domain of pp32 is required for the interaction with histone chaperone, TAF-Ibeta.Biol Pharm Bull. 2006 Dec;29(12):2395-8. doi: 10.1248/bpb.29.2395. Biol Pharm Bull. 2006. PMID: 17142970
-
Negative glucocorticoid receptor response elements and their role in glucocorticoid action.Curr Pharm Des. 2004;10(23):2807-16. doi: 10.2174/1381612043383601. Curr Pharm Des. 2004. PMID: 15379669 Review.
-
How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering.Mol Cell Endocrinol. 2013 Nov 5;380(1-2):41-54. doi: 10.1016/j.mce.2012.12.014. Epub 2012 Dec 23. Mol Cell Endocrinol. 2013. PMID: 23267834 Review.
Cited by
-
Acetylation-mediated epigenetic regulation of glucocorticoid receptor activity: circadian rhythm-associated alterations of glucocorticoid actions in target tissues.Mol Cell Endocrinol. 2011 Apr 10;336(1-2):23-30. doi: 10.1016/j.mce.2010.12.001. Epub 2010 Dec 10. Mol Cell Endocrinol. 2011. PMID: 21146585 Free PMC article.
-
NUP214 in Leukemia: It's More than Transport.Cells. 2019 Jan 21;8(1):76. doi: 10.3390/cells8010076. Cells. 2019. PMID: 30669574 Free PMC article. Review.
-
Oncoprotein SET dynamically regulates cellular stress response through nucleocytoplasmic transport in breast cancer.Cell Biol Toxicol. 2023 Aug;39(4):1795-1814. doi: 10.1007/s10565-022-09784-4. Epub 2022 Dec 19. Cell Biol Toxicol. 2023. PMID: 36534342
-
The SET oncoprotein promotes estrogen-induced transcription by facilitating establishment of active chromatin.Proc Natl Acad Sci U S A. 2023 Feb 21;120(8):e2206878120. doi: 10.1073/pnas.2206878120. Epub 2023 Feb 15. Proc Natl Acad Sci U S A. 2023. PMID: 36791099 Free PMC article.
-
The next decade of SET: from an oncoprotein to beyond.J Mol Cell Biol. 2024 Jul 1;16(1):mjad082. doi: 10.1093/jmcb/mjad082. J Mol Cell Biol. 2024. PMID: 38157418 Free PMC article. Review.
References
-
- Chrousos GP. The hypothalamic-pituitary-adrenal axis and immune-mediated inflammation. N Engl J Med. 1995;332:1351–1362. - PubMed
-
- Chrousos GP. Glucocorticoid therapy. In: Felig P, Frohman LA, editors. Endocrinology & Metabolism. McGraw-Hill; New York: 2001. pp. 609–632.
-
- Kino T, De Martino MU, Charmandari E, Mirani M, Chrousos GP. Tissue glucocorticoid resistance/hypersensitivity syndromes. J Steroid Biochem Mol Biol. 2003;85:457–467. - PubMed
-
- Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11(Suppl 1):S45–55. - PubMed
-
- Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid receptors and associated diseases. Essays Biochem. 2004;40:137–155. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

