HU binds and folds single-stranded DNA
- PMID: 18096614
- PMCID: PMC2241890
- DOI: 10.1093/nar/gkm667
HU binds and folds single-stranded DNA
Abstract
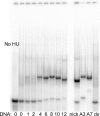
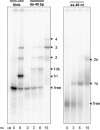
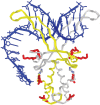
The nucleoid-associated protein HU plays an important role in bacterial nucleoid organization and is involved in numerous processes including transposition, recombination and DNA repair. We show here that HU binds specifically DNA containing mismatched region longer than 3 bp as well as DNA bulges. HU binds single-stranded DNA (ssDNA) in a binding mode that is reminiscent but different from earlier reported specific HU interactions with double-helical DNA lesions. An HU dimer requires 24 nt of ssDNA for initial binding, and 12 nt of ssDNA for each additional dimer binding. In the presence of equimolar amounts of HU dimer and DNA, the ssDNA molecule forms an U-loop (hairpin-like) around the protein, providing contacts with both sides of the HU body. This mode differs from the binding of the single-strand-binding protein (SSB) to ssDNA: in sharp contrast to SSB, HU binds ssDNA non-cooperatively and does not destabilize double-helical DNA. Furthermore HU has a strong preference for poly(dG), while binding to poly(dA) is the weakest. HU binding to ssDNA is probably important for its capacity to cover and protect bacterial DNA both intact and carrying lesions.
Figures



References
-
- Rouviere-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. - PubMed
-
- Rouviere-Yaniv J. Localization of the HU protein on the Escherichia coli nucleoid. Cold Spring Harb. Symp. Quant. Biol. 1978;42:439–447. - PubMed
-
- Wery M, Woldringh CL, Rouviere-Yaniv J. HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie. 2001;83:193–200. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

