Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions
- PMID: 18096747
- PMCID: PMC2203622
- DOI: 10.1101/gr.6887408
Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions
Abstract
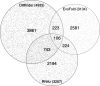
Recent computational scans for non-coding RNAs (ncRNAs) in multiple organisms have relied on existing multiple sequence alignments. However, as sequence similarity drops, a key signal of RNA structure--frequent compensating base changes--is increasingly likely to cause sequence-based alignment methods to misalign, or even refuse to align, homologous ncRNAs, consequently obscuring that structural signal. We have used CMfinder, a structure-oriented local alignment tool, to search the ENCODE regions of vertebrate multiple alignments. In agreement with other studies, we find a large number of potential RNA structures in the ENCODE regions. We report 6587 candidate regions with an estimated false-positive rate of 50%. More intriguingly, many of these candidates may be better represented by alignments taking the RNA secondary structure into account than those based on primary sequence alone, often quite dramatically. For example, approximately one-quarter of our predicted motifs show revisions in >50% of their aligned positions. Furthermore, our results are strongly complementary to those discovered by sequence-alignment-based approaches--84% of our candidates are not covered by Washietl et al., increasing the number of ncRNA candidates in the ENCODE region by 32%. In a group of 11 ncRNA candidates that were tested by RT-PCR, 10 were confirmed to be present as RNA transcripts in human tissue, and most show evidence of significant differential expression across tissues. Our results broadly suggest caution in any analysis relying on multiple sequence alignments in less well-conserved regions, clearly support growing appreciation for the biological significance of ncRNAs, and strongly support the argument for considering RNA structure directly in any searches for these elements.
Figures


References
-
- Bertone P., Stoc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stoc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
-
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning mulitple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. - PMC - PubMed
-
- Cavaille J., Vitali P., Basyuk E., Huttenhofer A., Bachellerie J.P., Vitali P., Basyuk E., Huttenhofer A., Bachellerie J.P., Basyuk E., Huttenhofer A., Bachellerie J.P., Huttenhofer A., Bachellerie J.P., Bachellerie J.P. A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem. 2001;276:26374–26383. - PubMed
-
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. - PubMed
-
- Costa F.F. Non-coding RNAs: New players in eukaryotic biology. Gene. 2005;357:83–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
