Mechanisms of resistance to an amino acid antibiotic that targets translation
- PMID: 18154269
- PMCID: PMC2386896
- DOI: 10.1021/cb7002253
Mechanisms of resistance to an amino acid antibiotic that targets translation
Erratum in
- ACS Chem Biol. 2008 Feb 15;3(2):130
Abstract
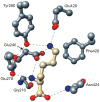
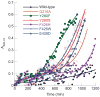
Structural and functional diversity among the aminoacyl-tRNA synthetases prevent infiltration of the genetic code by noncognate amino acids. To explore whether these same features distinguish the synthetases as potential sources of resistance against antibiotic amino acid analogues, we investigated bacterial growth inhibition by S-(2-aminoethyl)-L-cysteine (AEC). Wild-type lysyl-tRNA synthetase (LysRS) and a series of active site variants were screened for their ability to restore growth of an Escherichia coli LysRS null strain at increasing concentrations of AEC. While wild-type E. coli growth is completely inhibited at 5 microM AEC, two LysRS variants, Y280F and F426W, provided substantial resistance and allowed E. coli to grow in the presence of up to 1 mM AEC. Elevated resistance did not reflect changes in the kinetics of amino acid activation or tRNA (Lys) aminoacylation, which showed at best 4-6-fold improvements, but instead correlated with the binding affinity for AEC, which was decreased approximately 50-fold in the LysRS variants. In addition to changes in LysRS, AEC resistance has also been attributed to mutations in the L box riboswitch, which regulates expression of the lysC gene, encoding aspartokinase. The Y280F and F426W LysRS mutants contained wild-type L box riboswitches that responded normally to AEC in vitro, indicating that LysRS is the primary cellular target of this antibiotic. These findings suggest that the AEC resistance conferred by L box mutations is an indirect effect resulting from derepression of lysC expression and increased cellular pools of lysine, which results in more effective competition with AEC for binding to LysRS.
Figures


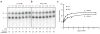
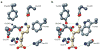
Similar articles
-
Nonorthologous replacement of lysyl-tRNA synthetase prevents addition of lysine analogues to the genetic code.Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):14351-6. doi: 10.1073/pnas.2036253100. Epub 2003 Nov 17. Proc Natl Acad Sci U S A. 2003. PMID: 14623972 Free PMC article.
-
Divergence in noncognate amino acid recognition between class I and class II lysyl-tRNA synthetases.J Biol Chem. 2004 Apr 23;279(17):17707-14. doi: 10.1074/jbc.M313665200. Epub 2004 Jan 27. J Biol Chem. 2004. PMID: 14747465
-
A single substitution in the motif 1 of Escherichia coli lysyl-tRNA synthetase induces cooperativity toward amino acid binding.Biochemistry. 1995 Jun 27;34(25):8180-9. doi: 10.1021/bi00025a025. Biochemistry. 1995. PMID: 7794932
-
Lysyl-tRNA synthetase.Biol Chem Hoppe Seyler. 1995 Aug;376(8):451-72. doi: 10.1515/bchm3.1995.376.8.451. Biol Chem Hoppe Seyler. 1995. PMID: 7576245 Review.
-
Structural analyses of the malaria parasite aminoacyl-tRNA synthetases provide new avenues for antimalarial drug discovery.Protein Sci. 2021 Sep;30(9):1793-1803. doi: 10.1002/pro.4148. Protein Sci. 2021. PMID: 34184352 Free PMC article. Review.
Cited by
-
Crystal structure of the lysine riboswitch regulatory mRNA element.J Biol Chem. 2008 Aug 15;283(33):22347-51. doi: 10.1074/jbc.C800120200. Epub 2008 Jul 1. J Biol Chem. 2008. PMID: 18593706 Free PMC article.
-
Prospects for riboswitch discovery and analysis.Mol Cell. 2011 Sep 16;43(6):867-79. doi: 10.1016/j.molcel.2011.08.024. Mol Cell. 2011. PMID: 21925376 Free PMC article. Review.
-
Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs.Acc Chem Res. 2011 Dec 20;44(12):1329-38. doi: 10.1021/ar200039b. Epub 2011 May 26. Acc Chem Res. 2011. PMID: 21615107 Free PMC article. Review.
-
Design and antimicrobial action of purine analogues that bind Guanine riboswitches.ACS Chem Biol. 2009 Nov 20;4(11):915-27. doi: 10.1021/cb900146k. ACS Chem Biol. 2009. PMID: 19739679 Free PMC article.
-
Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs.Structure. 2009 Jun 10;17(6):857-68. doi: 10.1016/j.str.2009.04.009. Structure. 2009. PMID: 19523903 Free PMC article.
References
-
- Ibba M, Söll D. Science. Vol. 286. 1999. Quality control mechanisms during translation; pp. 1893–1897. - PubMed
-
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, Rao CV, Tournev I, Gondim FA, D’Hooghe M, Van Gerwen V, Callaerts P, Van Den BL, Timmermans JP, Robberecht W, Gettemans J, Thevelein JM, De Jonghe P, Kremensky I, Tim-merman V. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. - PubMed
-
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. - PubMed
-
- Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13:1091–1100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

