Prediction of salt and mutational effects on the association rate of U1A protein and U1 small nuclear RNA stem/loop II
- PMID: 18154282
- PMCID: PMC3526768
- DOI: 10.1021/jp075919k
Prediction of salt and mutational effects on the association rate of U1A protein and U1 small nuclear RNA stem/loop II
Abstract
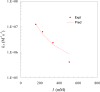
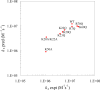
We have developed a computational approach for predicting protein-protein association rates (Alsallaq and Zhou, Structure 2007, 15, 215). Here we expand the range of applicability of this approach to protein-RNA binding and report the first results for protein-RNA binding rates predicted from atomistic modeling. The system studied is the U1A protein and stem/loop II of the U1 small nuclear RNA. Experimentally it was observed that the binding rate is significantly reduced by increasing salt concentration while the dissociation changes little with salt concentration, and charges distant from the binding site make marginal contribution to the binding rate. These observations are rationalized. Moreover, predicted effects of salt and charge mutations are found to be in quantitative agreement with experimental results.
Figures



Similar articles
-
Investigating the binding specificity of U1A-RNA by computational mutagenesis.J Mol Biol. 2000 Jan 7;295(1):1-6. doi: 10.1006/jmbi.1999.3319. J Mol Biol. 2000. PMID: 10623503
-
Molecular dynamics simulations of the complex between human U1A protein and hairpin II of U1 small nuclear RNA and of free RNA in solution.Biophys J. 1999 Sep;77(3):1284-305. doi: 10.1016/S0006-3495(99)76979-1. Biophys J. 1999. PMID: 10465742 Free PMC article.
-
Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA.J Biol Chem. 2001 Jun 15;276(24):21476-81. doi: 10.1074/jbc.M101624200. Epub 2001 Apr 10. J Biol Chem. 2001. PMID: 11297556
-
RNA-protein interactions. Diverse modes of recognition.Curr Biol. 1995 Mar 1;5(3):249-51. doi: 10.1016/s0960-9822(95)00051-0. Curr Biol. 1995. PMID: 7540101 Review.
-
Model of the interaction between the UI A small nuclear ribonucleoprotein and UI small nuclear RNA stem/loop II.Biochem Soc Trans. 1993 Aug;21 ( Pt 3)(3):605-9. doi: 10.1042/bst0210605. Biochem Soc Trans. 1993. PMID: 8224475 Review. No abstract available.
Cited by
-
On the Dielectric Boundary in Poisson-Boltzmann Calculations.J Chem Theory Comput. 2008 Mar;4(3):507-514. doi: 10.1021/ct700319x. Epub 2008 Feb 21. J Chem Theory Comput. 2008. PMID: 23304097 Free PMC article.
-
Energetic evaluation of binding modes in the C3d and Factor H (CCP 19-20) complex.Protein Sci. 2015 May;24(5):789-802. doi: 10.1002/pro.2650. Epub 2015 Mar 11. Protein Sci. 2015. PMID: 25628052 Free PMC article.
-
Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation.Chem Rev. 2018 Feb 28;118(4):1691-1741. doi: 10.1021/acs.chemrev.7b00305. Epub 2018 Jan 10. Chem Rev. 2018. PMID: 29319301 Free PMC article. Review.
-
Rationalizing 5000-fold differences in receptor-binding rate constants of four cytokines.Biophys J. 2011 Sep 7;101(5):1175-83. doi: 10.1016/j.bpj.2011.06.056. Biophys J. 2011. PMID: 21889455 Free PMC article.
-
Automated prediction of protein association rate constants.Structure. 2011 Dec 7;19(12):1744-51. doi: 10.1016/j.str.2011.10.015. Structure. 2011. PMID: 22153497 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

