The histamine N-methyltransferase T105I polymorphism affects active site structure and dynamics
- PMID: 18154359
- PMCID: PMC2905460
- DOI: 10.1021/bi701737f
The histamine N-methyltransferase T105I polymorphism affects active site structure and dynamics
Abstract
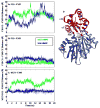
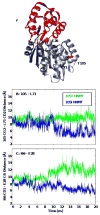
Histamine N-methyltransferase (HNMT) is the primary enzyme responsible for inactivating histamine in the mammalian brain. The human HNMT gene contains a common threonine-isoleucine polymorphism at residue 105, distal from the active site. The 105I variant has decreased activity and lower protein levels than the 105T protein. Crystal structures of both variants have been determined but reveal little regarding how the T105I polymorphism affects activity. We performed molecular dynamics simulations for both 105T and 105I at 37 degrees C to explore the structural and dynamic consequences of the polymorphism. The simulations indicate that replacing Thr with the larger Ile residue leads to greater burial of residue 105 and heightened intramolecular interactions between residue 105 and residues within helix alpha3 and strand beta3. This altered, tighter packing is translated to the active site, resulting in the reorientation of several cosubstrate-binding residues. The simulations also show that the hydrophobic histamine-binding domain in both proteins undergoes a large-scale breathing motion that exposes key catalytic residues and lowers the hydrophobicity of the substrate-binding site.
Figures


 for α-helices, and
for α-helices, and  for β-strands, and are colored to match the SAM- and histamine-binding domains shown in Figure 1. * = residue 105.
for β-strands, and are colored to match the SAM- and histamine-binding domains shown in Figure 1. * = residue 105.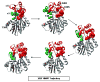

Similar articles
-
Histamine N-Methyltransferase in the Brain.Int J Mol Sci. 2019 Feb 10;20(3):737. doi: 10.3390/ijms20030737. Int J Mol Sci. 2019. PMID: 30744146 Free PMC article. Review.
-
Theoretical 3D model of histamine N-methyltransferase: insights into the effects of a genetic polymorphism on enzymatic activity and thermal stability.Biochem Biophys Res Commun. 2001 Sep 14;287(1):204-8. doi: 10.1006/bbrc.2001.5570. Biochem Biophys Res Commun. 2001. PMID: 11549275
-
Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.Structure. 2001 Sep;9(9):837-49. doi: 10.1016/s0969-2126(01)00643-8. Structure. 2001. PMID: 11566133 Free PMC article.
-
Elucidating the impact of S-adenosylmethionine and histamine binding on N-methyltransferase conformational dynamics: Insights from an in silico study.J Mol Graph Model. 2025 May;136:108961. doi: 10.1016/j.jmgm.2025.108961. Epub 2025 Jan 24. J Mol Graph Model. 2025. PMID: 39879846
-
The Thr105Ile Variant (rs11558538) of the Histamine N-methyltransferase Gene may be associated with Reduced Risk of Parkinson Disease: A Meta-analysis.Genet Test Mol Biomarkers. 2022 Nov;26(11):543-549. doi: 10.1089/gtmb.2021.0299. Epub 2022 Nov 15. Genet Test Mol Biomarkers. 2022. PMID: 36378841
Cited by
-
The diamine oxidase gene is associated with hypersensitivity response to non-steroidal anti-inflammatory drugs.PLoS One. 2012;7(11):e47571. doi: 10.1371/journal.pone.0047571. Epub 2012 Nov 12. PLoS One. 2012. PMID: 23152756 Free PMC article.
-
Polymorphisms and disease: hotspots of inactivation in methyltransferases.Trends Biochem Sci. 2010 Oct;35(10):531-8. doi: 10.1016/j.tibs.2010.03.007. Epub 2010 Apr 9. Trends Biochem Sci. 2010. PMID: 20382027 Free PMC article.
-
Histamine N-Methyltransferase in the Brain.Int J Mol Sci. 2019 Feb 10;20(3):737. doi: 10.3390/ijms20030737. Int J Mol Sci. 2019. PMID: 30744146 Free PMC article. Review.
-
Histamine N-methyltransferase Thr105Ile is not associated with Parkinson's disease or essential tremor.Parkinsonism Relat Disord. 2010 Feb;16(2):112-4. doi: 10.1016/j.parkreldis.2009.08.011. Epub 2009 Sep 20. Parkinsonism Relat Disord. 2010. PMID: 19773194 Free PMC article.
-
Human variability in isoform-specific UDP-glucuronosyltransferases: markers of acute and chronic exposure, polymorphisms and uncertainty factors.Arch Toxicol. 2020 Aug;94(8):2637-2661. doi: 10.1007/s00204-020-02765-8. Epub 2020 May 15. Arch Toxicol. 2020. PMID: 32415340 Free PMC article.
References
-
- Rosenwasser L. New insights into the pathophysiology of allergic rhinitis. Allergy Asthma Proc. 2007;28:10–5. - PubMed
-
- Zhang M, Thurmond RL, Dunford PJ. The histamine H(4) receptor: a novel modulator of inflammatory and immune disorders. Pharmacol Ther. 2007;113:594–606. - PubMed
-
- Schubert ML. Regulation of gastric acid secretion. Curr Opin Gastroenterol. 1999;15:457. - PubMed
-
- Chen D, Friis-Hansen L, Hakanson R, Zhao CM. Genetic dissection of the signaling pathways that control gastric acid secretion. Inflammopharmacology. 2005;13:201–7. - PubMed
-
- Bacciottini L, Passani MB, Mannaioni PF, Blandina P. Interactions between histaminergic and cholinergic systems in learning and memory. Behav Brain Res. 2001;124:183–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

