Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants
- PMID: 18154671
- PMCID: PMC2234419
- DOI: 10.1186/1471-2148-7-248
Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants
Abstract
Background: Some of the most difficult phylogenetic questions in evolutionary biology involve identification of the free-living relatives of parasitic organisms, particularly those of parasitic flowering plants. Consequently, the number of origins of parasitism and the phylogenetic distribution of the heterotrophic lifestyle among angiosperm lineages is unclear.
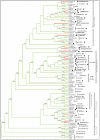
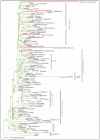
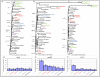
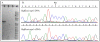
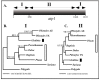
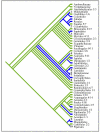
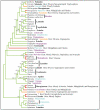
Results: Here we report the results of a phylogenetic analysis of 102 species of seed plants designed to infer the position of all haustorial parasitic angiosperm lineages using three mitochondrial genes: atp1, coxI, and matR. Overall, the mtDNA phylogeny agrees with independent studies in terms of non-parasitic plant relationships and reveals at least 11 independent origins of parasitism in angiosperms, eight of which consist entirely of holoparasitic species that lack photosynthetic ability. From these results, it can be inferred that modern-day parasites have disproportionately evolved in certain lineages and that the endoparasitic habit has arisen by convergence in four clades. In addition, reduced taxon, single gene analyses revealed multiple horizontal transfers of atp1 from host to parasite lineage, suggesting that parasites may be important vectors of horizontal gene transfer in angiosperms. Furthermore, in Pilostyles we show evidence for a recent host-to-parasite atp1 transfer based on a chimeric gene sequence that indicates multiple historical xenologous gene acquisitions have occurred in this endoparasite. Finally, the phylogenetic relationships inferred for parasites indicate that the origins of parasitism in angiosperms are strongly correlated with horizontal acquisitions of the invasive coxI group I intron.
Conclusion: Collectively, these results indicate that the parasitic lifestyle has arisen repeatedly in angiosperm evolutionary history and results in increasing parasite genomic chimerism over time.
Figures
References
-
- Combes C. Parasitism: the Ecology and Evolution of Intimate Interactions. Chicago: The University of Chicago Press; 2001.
-
- Bush AO, Fernandez JC, Esch GW, Seed JR. Parasitism: The Diversity and Ecology of Animal Parasites. Cambridge: Cambridge University Press; 2001.
-
- Kuijt J. The Biology of Parasitic Flowering Plants. Berkeley: University of California Press; 1969.
-
- Nais J. Rafflesia of the World. Kota Kinabalu: Natural History Publications; 2001.
-
- Nickrent DL, Duff RJ, Colwell AE, Wolfe AD, Young ND, Steiner KE, dePamphilis CW. Molecular phylogenetic and evolutionary studies of parasitic plants. In: Soltis DE, Soltis PS, Doyle JJ, editor. Molecular Systematics of Plants II DNA Sequencing. Boston, Massachusetts: Kluwer Academic; 1998. pp. 211–241.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

