Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling
- PMID: 18156209
- PMCID: PMC2189623
- DOI: 10.2353/ajpath.2008.070294
Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling
Abstract
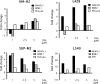
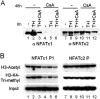
The nuclear factor of activated T cell 1 (Nfatc1) locus is a common insertion site for murine tumorigenic retroviruses, suggesting a role of transcription factor NFATc1 in lymphomagenesis. Although NFATc1 is expressed in most human primary lymphocytes and mature human T- and B-cell neoplasms, we show by histochemical stainings that NFATc1 expression is suppressed in anaplastic large cell lymphomas and classical Hodgkin's lymphomas (HLs). In HL cell lines, NFATc1 silencing correlated with a decrease in histone H3 acetylation, H3-K4 trimethylation, and Sp1 factor binding but with an increase in HP1 binding to the NFATC1 P1 promoter. Together with DNA hypermethylation of the NFATC1 P1 promoter, which we detected in all anaplastic large cell lymphoma and many HL lines, these observations reflect typical signs of transcriptional silencing. In several lymphoma lines, methylation of NFATC1 promoter DNA resulted in a "window of hypomethylation," which is flanked by Sp1-binding sites. Together with the under-representation of Sp1 at the NFATC1 P1 promoter in HL cells, this suggests that Sp1 factors can protect P1 DNA methylation in a directional manner. Blocking immunoreceptor signaling led to NFATC1 P1 promoter silencing and to a decrease in H3 acetylation and H3-K4 methylation but not DNA methylation. This shows that histone modifications precede the DNA methylation in NFATC1 promoter silencing.
Figures




References
-
- Serfling E, Berberich-Siebelt F, Chuvpilo S, Jankevics E, Klein-Hessling S, Twardzik T, Avots A. The role of NF-AT transcription factors in T cell activation and differentiation. Biochim Biophys Acta. 2000;1498:1–18. - PubMed
-
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. - PubMed
-
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. - PubMed
-
- Baksh S, Widlund HR, Frazer-Abel AA, Du J, Fosmire S, Fisher DE, DeCaprio JA, Modiano JF, Burakoff SJ. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol Cell. 2002;10:1071–1081. - PubMed
-
- Latinis KM, Norian LA, Eliason SL, Koretzky GA. Two NFAT transcription factor binding sites participate in the regulation of CD95 (Fas) ligand expression in activated human T cells. J Biol Chem. 1997;272:31427–31434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

