3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula
- PMID: 18156218
- PMCID: PMC2217646
- DOI: 10.1105/tpc.107.053975
3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula
Abstract
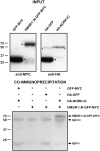
NORK in legumes encodes a receptor-like kinase that is required for Nod factor signaling and root nodule development. Using Medicago truncatula NORK as bait in a yeast two-hybrid assay, we identified 3-hydroxy-3-methylglutaryl CoA reductase 1 (Mt HMGR1) as a NORK interacting partner. HMGR1 belongs to a multigene family in M. truncatula, and different HMGR isoforms are key enzymes in the mevalonate biosynthetic pathway leading to the production of a diverse array of isoprenoid compounds. Testing other HMGR members revealed a specific interaction between NORK and HMGR1. Mutagenesis and deletion analysis showed that this interaction requires the cytosolic active kinase domain of NORK and the cytosolic catalytic domain of HMGR1. NORK homologs from Lotus japonicus and Sesbania rostrata also interacted with Mt HMGR1, but homologous nonsymbiotic kinases of M. truncatula did not. Pharmacological inhibition of HMGR activities decreased nodule number and delayed nodulation, supporting the importance of the mevalonate pathway in symbiotic development. Decreasing HMGR1 expression in M. truncatula transgenic roots by RNA interference led to a dramatic decrease in nodulation, confirming that HMGR1 is essential for nodule development. Recruitment of HMGR1 by NORK could be required for production of specific isoprenoid compounds, such as cytokinins, phytosteroids, or isoprenoid moieties involved in modification of signaling proteins.
Figures





References
-
- Ané, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. - PubMed
-
- Batut, J., Andersson, S.G., and O'Callaghan, D. (2004). The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2 933–945. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

