In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections
- PMID: 18156255
- PMCID: PMC2293235
- DOI: 10.1128/JB.01581-07
In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections
Abstract
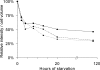
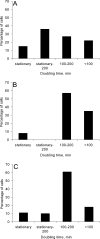
The growth dynamics of bacterial pathogens within infected hosts are a fundamental but poorly understood feature of most infections. We have focused on the in situ distribution and growth characteristics of two prevailing and transmissible Pseudomonas aeruginosa clones that have caused chronic lung infections in cystic fibrosis (CF) patients for more than 20 years. We used fluorescence in situ hybridization (FISH) directly on sputum specimens to examine the spatial distribution of the infecting P. aeruginosa cells. Mucoid variants were present in sputum as cell clusters surrounded by an extracellular matrix, whereas nonmucoid variants were present mainly as dispersed cells. To obtain estimates of the growth rates of P. aeruginosa in CF lungs, we used quantitative FISH to indirectly measure growth rates of bacteria in sputum samples (reflecting the in vivo lung conditions). The concentration of rRNA in bacteria isolated from sputa was measured and correlated with the rRNA contents of the same bacteria growing in vitro at defined rates. The results showed that most cells were actively growing with doubling times of between 100 and 200 min, with some growing even faster. Only a small stationary-phase subpopulation seemed to be present in sputa. This was found for both mucoid and nonmucoid variants despite their different organizations in sputum. The results suggest that the bacterial population may be confronted with selection forces that favor optimized growth activities. This scenario constitutes a new perspective on the adaptation and evolution of P. aeruginosa during chronic infections in CF patients in particular and on long-term infections in general.
Figures



References
-
- Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2489-493. - PubMed
-
- Baltimore, R. S., C. D. Christie, and G. J. Smith. 1989. Immunohistopathologic localization of Pseudomonas aeruginosa in lungs from patients with cystic fibrosis. Implications for the pathogenesis of progressive lung deterioration. Am. Rev. Respir. Dis. 1401650-1661. - PubMed
-
- Barth, A. L., and T. L. Pitt. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45110-119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

